Identification of a regulatory pathway inhibiting adipogenesis via RSPO2
- PMID: 35027768
- PMCID: PMC8803606
- DOI: 10.1038/s42255-021-00509-1
Identification of a regulatory pathway inhibiting adipogenesis via RSPO2
Abstract
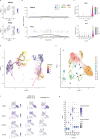
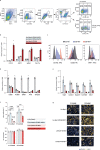
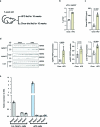
Healthy adipose tissue remodeling depends on the balance between de novo adipogenesis from adipogenic progenitor cells and the hypertrophy of adipocytes. De novo adipogenesis has been shown to promote healthy adipose tissue expansion, which confers protection from obesity-associated insulin resistance. Here, we define the role and trajectory of different adipogenic precursor subpopulations and further delineate the mechanism and cellular trajectory of adipogenesis, using single-cell RNA-sequencing datasets of murine adipogenic precursors. We identify Rspo2 as a functional regulator of adipogenesis, which is secreted by a subset of CD142+ cells to inhibit maturation of early progenitors through the receptor Lgr4. Increased circulating RSPO2 in mice leads to adipose tissue hypertrophy and insulin resistance and increased RSPO2 levels in male obese individuals correlate with impaired glucose homeostasis. Taken together, these findings identify a complex cellular crosstalk that inhibits adipogenesis and impairs adipose tissue homeostasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














References
-
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. - PubMed
-
- Muller S, Kulenkampff E, Wolfrum C. Adipose tissue stem cells. Handb. Exp. Pharmacol. 2016;233:251–263. - PubMed
-
- Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019;20:242–258. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

