Altered glycosylation of several metastasis-associated glycoproteins with terminal GalNAc defines the highly invasive cancer cell phenotype
- PMID: 35028012
- PMCID: PMC8751650
- DOI: 10.18632/oncotarget.28167
Altered glycosylation of several metastasis-associated glycoproteins with terminal GalNAc defines the highly invasive cancer cell phenotype
Abstract
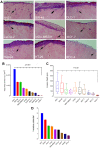
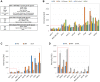
Several distinct metastasis-associated glycosylation changes have been shown to promote cancer cell invasion and metastasis, the main cause of death of cancer patients. However, it is unclear whether their presence reflects cell- or tissue-specific variations for metastasis, or species needed to drive different phases of the metastatic cascade. To address this issue from a different perspective, we investigated here whether different cancer cell lines share any glycotopes that are common and important for their invasive phenotype. By using lectin microarray glycan profiling and an established myoma tissue-based 3D invasion assay, we identified a single glycotope recognized by Helix Pomatia agglutinin (HPA), whose expression level in different cancer cells correlated significantly with their invasive potential. Lectin pull-down assay and LC-MS/MS analysis in highly- (A431 and SW-48) and poorly invasive (HepG2 and RCC4) cancer cells revealed ~85 glycoproteins of which several metastasis-promoting members of the integrin family of cell adhesion receptors, the epidermal growth factor receptor (EGFR) and the matrix metalloproteinase-14 (MMP-14) were among the abundant ones. Moreover, we showed that the level of the GalNAc glycotope in MMP-14, EGFR, αV-, β1- and β4 integrin in highly and poorly invasive cancer cells correlated positively with their invasive potential. Collectively, our findings suggest that altered glycosylation of several metastasis-associated glycoproteins with terminal GalNAc drives the highly invasive cancer cell phenotype.
Keywords: Helix Pomatia agglutinin (HPA); cancer; glycocode; glycosylation; invasion; terminal GalNAc.
Copyright: © 2022 Khosrowabadi et al.
Conflict of interest statement
CONFLICTS OF INTEREST Authors have no conflicts of interest to declare.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

