Multiomic analysis identifies CPT1A as a potential therapeutic target in platinum-refractory, high-grade serous ovarian cancer
- PMID: 35028612
- PMCID: PMC8714940
- DOI: 10.1016/j.xcrm.2021.100471
Multiomic analysis identifies CPT1A as a potential therapeutic target in platinum-refractory, high-grade serous ovarian cancer
Abstract
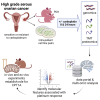
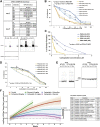
Resistance to platinum compounds is a major determinant of patient survival in high-grade serous ovarian cancer (HGSOC). To understand mechanisms of platinum resistance and identify potential therapeutic targets in resistant HGSOC, we generated a data resource composed of dynamic (±carboplatin) protein, post-translational modification, and RNA sequencing (RNA-seq) profiles from intra-patient cell line pairs derived from 3 HGSOC patients before and after acquiring platinum resistance. These profiles reveal extensive responses to carboplatin that differ between sensitive and resistant cells. Higher fatty acid oxidation (FAO) pathway expression is associated with platinum resistance, and both pharmacologic inhibition and CRISPR knockout of carnitine palmitoyltransferase 1A (CPT1A), which represents a rate limiting step of FAO, sensitize HGSOC cells to platinum. The results are further validated in patient-derived xenograft models, indicating that CPT1A is a candidate therapeutic target to overcome platinum resistance. All multiomic data can be queried via an intuitive gene-query user interface (https://sites.google.com/view/ptrc-cell-line).
Keywords: CPT1A; carboplatin; fatty acid oxidation; ovarian cancer; oxidative phosphorylation; proteogenomic; proteomic; reactive oxygen species; resistance.
© 2021 The Author(s).
Conflict of interest statement
M.J.B. has participated in advisory boards for Clovis, Astra Zeneca, and GSK-Tesaro. A.N.H. has a financial interest in the company Seattle Genetics.
Figures






References
-
- DeVita V.T., Rosenberg S.A., Lawrence T.S. Eleventh Edition. Wolters Kluwer; 2018. DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology (Cancer Principles and Practice of Oncology)
-
- Pascoe J.M., Roberts J.J. Interactions between mammalian cell DNA and inorganic platinum compounds. I. DNA interstrand cross-linking and cytotoxic properties of platinum(II) compounds. Biochem. Pharmacol. 1974;23:1359–1365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

