Down, then up: non-parallel genome size changes and a descending chromosome series in a recent radiation of the Australian allotetraploid plant species, Nicotiana section Suaveolentes (Solanaceae)
- PMID: 35029647
- PMCID: PMC9904355
- DOI: 10.1093/aob/mcac006
Down, then up: non-parallel genome size changes and a descending chromosome series in a recent radiation of the Australian allotetraploid plant species, Nicotiana section Suaveolentes (Solanaceae)
Abstract
Background and aims: The extent to which genome size and chromosome numbers evolve in concert is little understood, particularly after polyploidy (whole-genome duplication), when a genome returns to a diploid-like condition (diploidization). We study this phenomenon in 46 species of allotetraploid Nicotiana section Suaveolentes (Solanaceae), which formed <6 million years ago and radiated in the arid centre of Australia.
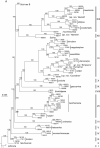
Methods: We analysed newly assessed genome sizes and chromosome numbers within the context of a restriction site-associated nuclear DNA (RADseq) phylogenetic framework.
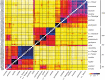
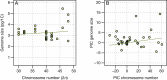
Key results: RADseq generated a well-supported phylogenetic tree, in which multiple accessions from each species formed unique genetic clusters. Chromosome numbers and genome sizes vary from n = 2x = 15 to 24 and 2.7 to 5.8 pg/1C nucleus, respectively. Decreases in both genome size and chromosome number occur, although neither consistently nor in parallel. Species with the lowest chromosome numbers (n = 15-18) do not possess the smallest genome sizes and, although N. heterantha has retained the ancestral chromosome complement, n = 2x = 24, it nonetheless has the smallest genome size, even smaller than that of the modern representatives of ancestral diploids.
Conclusions: The results indicate that decreases in genome size and chromosome number occur in parallel down to a chromosome number threshold, n = 20, below which genome size increases, a phenomenon potentially explained by decreasing rates of recombination over fewer chromosomes. We hypothesize that, more generally in plants, major decreases in genome size post-polyploidization take place while chromosome numbers are still high because in these stages elimination of retrotransposons and other repetitive elements is more efficient. Once such major genome size change has been accomplished, then dysploid chromosome reductions take place to reorganize these smaller genomes, producing species with small genomes and low chromosome numbers such as those observed in many annual angiosperms, including Arabidopsis.
Keywords: Nicotiana benthamiana; Nicotiana sect. Suaveolentes; Allotetraploid evolution; Australian endemics; C-value; Solanaceae; WGD; diploidization; dysploidy; epigenetics; model organism; polyploidy.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures



References
-
- Bally J, Marks C, Jung H, et al. 2021. Nicotiana paulineana, a new Australian species in Nicotiana section Suaveolentes. Australian Systematic Botany 34: 477–484.
-
- Blomberg SP, Lefevre JG, Wells JA, Waterhouse M. 2012. Independent contrasts and PGLS regression estimators are equivalent. Systematic Biology 61: 382–391. - PubMed
-
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB. 2012. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant–microbe biology research. Molecular Plant-Microbe Interactions 25: 1523–1530. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

