N protein-based ultrasensitive SARS-CoV-2 antibody detection in seconds via 3D nanoprinted, microarchitected array electrodes
- PMID: 35032037
- PMCID: PMC9015463
- DOI: 10.1002/jmv.27591
N protein-based ultrasensitive SARS-CoV-2 antibody detection in seconds via 3D nanoprinted, microarchitected array electrodes
Abstract
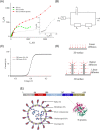
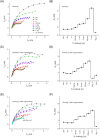
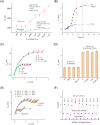
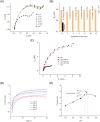
Rapid detection of antibodies to SARS-CoV-2 is critical for COVID-19 diagnostics, epidemiological research, and studies related to vaccine evaluation. It is known that the nucleocapsid (N) is the most abundant protein of SARS-CoV-2 and can serve as an excellent biomarker due to its strong immunogenicity. This paper reports a rapid and ultrasensitive 3D biosensor for quantification of COVID-19 antibodies in seconds via electrochemical transduction. This sensor consists of an array of three-dimensional micro-length-scale electrode architecture that is fabricated by aerosol jet 3D printing, which is an additive manufacturing technique. The micropillar array is coated with N proteins via an intermediate layer of nano-graphene and is integrated into a microfluidic channel to complete an electrochemical cell that uses antibody-antigen interaction to detect the antibodies to the N protein. Due to the structural innovation in the electrode geometry, the sensing is achieved in seconds, and the sensor shows an excellent limit of detection of 13 fm and an optimal detection range of 100 fm to 1 nm. Furthermore, the sensor can be regenerated at least 10 times, which reduces the cost per test. This work provides a powerful platform for rapid screening of antibodies to SARS-CoV-2 after infection or vaccination.
Keywords: 3D printing and functional sensors; N protein; SARS-CoV-2; antibody.
© 2022 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures

References
-
- Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS‐CoV‐2 variant of concern in South Africa. Nature. 2021;592(7854):438‐443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

