SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids
- PMID: 35032430
- PMCID: PMC8709832
- DOI: 10.1016/j.stem.2021.12.010
SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids
Abstract
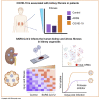
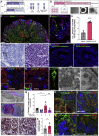
Kidney failure is frequently observed during and after COVID-19, but it remains elusive whether this is a direct effect of the virus. Here, we report that SARS-CoV-2 directly infects kidney cells and is associated with increased tubule-interstitial kidney fibrosis in patient autopsy samples. To study direct effects of the virus on the kidney independent of systemic effects of COVID-19, we infected human-induced pluripotent stem-cell-derived kidney organoids with SARS-CoV-2. Single-cell RNA sequencing indicated injury and dedifferentiation of infected cells with activation of profibrotic signaling pathways. Importantly, SARS-CoV-2 infection also led to increased collagen 1 protein expression in organoids. A SARS-CoV-2 protease inhibitor was able to ameliorate the infection of kidney cells by SARS-CoV-2. Our results suggest that SARS-CoV-2 can directly infect kidney cells and induce cell injury with subsequent fibrosis. These data could explain both acute kidney injury in COVID-19 patients and the development of chronic kidney disease in long COVID.
Keywords: COVID-19; SARS-CoV-2; chronic kidney disease; fibrosis; human iPSC kidney organoids; kidney injury; protease blocker.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
Potential SARS-CoV-2 kidney infection and paths to injury.Nat Rev Nephrol. 2022 May;18(5):275-276. doi: 10.1038/s41581-022-00551-6. Nat Rev Nephrol. 2022. PMID: 35190715 Free PMC article.
-
SARS-CoV-2 pirates the kidneys: A scar(y) story.Cell Metab. 2022 Mar 1;34(3):352-354. doi: 10.1016/j.cmet.2022.02.005. Cell Metab. 2022. PMID: 35235772 Free PMC article.
References
-
- Achdout H., Aimon A., Bar-David E., Barr H., Ben-Shmuel A., Bennett J., Bobby M.L., Brun J., Sarma B., et al. The COVID Moonshot Consortium COVID moonshot: open science discovery of SARS-CoV-2 main protease inhibitors by combining crowdsourcing, high-throughput experiments, computational simulations, and machine learning. BioRxiv. 2020 doi: 10.1101/2020.10.29.339317. - DOI
-
- Ajay A.K., Kim T.M., Ramirez-Gonzalez V., Park P.J., Frank D.A., Vaidya V.S. A bioinformatics approach identifies signal transducer and activator of transcription-3 and checkpoint kinase 1 as upstream regulators of kidney injury molecule-1 after kidney injury. J. Am. Soc. Nephrol. 2014;25:105–118. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

