Analysis of genetic variants in myeloproliferative neoplasms using a 22-gene next-generation sequencing panel
- PMID: 35033063
- PMCID: PMC8760696
- DOI: 10.1186/s12920-021-01145-0
Analysis of genetic variants in myeloproliferative neoplasms using a 22-gene next-generation sequencing panel
Abstract
Background: The Philadelphia (Ph)-negative myeloproliferative neoplasms (MPNs), namely essential thrombocythaemia (ET), polycythaemia vera (PV) and primary myelofibrosis (PMF), are a group of chronic clonal haematopoietic disorders that have the propensity to advance into bone marrow failure or acute myeloid leukaemia; often resulting in fatality. Although driver mutations have been identified in these MPNs, subtype-specific markers of the disease have yet to be discovered. Next-generation sequencing (NGS) technology can potentially improve the clinical management of MPNs by allowing for the simultaneous screening of many disease-associated genes.
Methods: The performance of a custom, in-house designed 22-gene NGS panel was technically validated using reference standards across two independent replicate runs. The panel was subsequently used to screen a total of 10 clinical MPN samples (ET n = 3, PV n = 3, PMF n = 4). The resulting NGS data was then analysed via a bioinformatics pipeline.
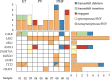
Results: The custom NGS panel had a detection limit of 1% variant allele frequency (VAF). A total of 20 unique variants with VAFs above 5% (4 of which were putatively novel variants with potential biological significance) and one pathogenic variant with a VAF of between 1 and 5% were identified across all of the clinical MPN samples. All single nucleotide variants with VAFs ≥ 15% were confirmed via Sanger sequencing.
Conclusions: The high fidelity of the NGS analysis and the identification of known and novel variants in this study cohort support its potential clinical utility in the management of MPNs. However, further optimisation is needed to avoid false negatives in regions with low sequencing coverage, especially for the detection of driver mutations in MPL.
Keywords: Bioinformatics, disease management; Gene; Mutation; Myeloproliferative neoplasm; Next-generation sequencing; Variant.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Cervantes F, Passamonti F, Barosi G. Life expectancy and prognostic factors in the classic BCR/ABL-negative myeloproliferative disorders. Leukemia. 2008;22(5):905–914. - PubMed
-
- Vannucchi AM, Barbui T, Cervantes F, Harrison C, Kiladjian JJ, Kroger N, et al. Philadelphia chromosome-negative chronic myeloproliferative neoplasms: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol Off J Eur Soc Med Oncol. 2015;26(Suppl 5):v85–v99. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, Jaffe E, Pileri S, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4. Lyon: International Agency for Research on Cancer (IARC) Press; 2017.
-
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

