Reduced and highly diverse peripheral HIV-1 reservoir in virally suppressed patients infected with non-B HIV-1 strains in Uganda
- PMID: 35033105
- PMCID: PMC8760765
- DOI: 10.1186/s12977-022-00587-3
Reduced and highly diverse peripheral HIV-1 reservoir in virally suppressed patients infected with non-B HIV-1 strains in Uganda
Abstract
Background: Our understanding of the peripheral human immunodeficiency virus type 1 (HIV-1) reservoir is strongly biased towards subtype B HIV-1 strains, with only limited information available from patients infected with non-B HIV-1 subtypes, which are the predominant viruses seen in low- and middle-income countries (LMIC) in Africa and Asia.
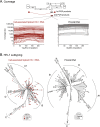
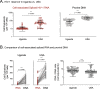
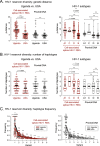
Results: In this study, blood samples were obtained from well-suppressed ART-experienced HIV-1 patients monitored in Uganda (n = 62) or the U.S. (n = 50), with plasma HIV-1 loads < 50 copies/ml and CD4+ T-cell counts > 300 cells/ml. The peripheral HIV-1 reservoir, i.e., cell-associated HIV-1 RNA and proviral DNA, was characterized using our novel deep sequencing-based EDITS assay. Ugandan patients were slightly younger (median age 43 vs 49 years) and had slightly lower CD4+ counts (508 vs 772 cells/ml) than U.S. individuals. All Ugandan patients were infected with non-B HIV-1 subtypes (31% A1, 64% D, or 5% C), while all U.S. individuals were infected with subtype B viruses. Unexpectedly, we observed a significantly larger peripheral inducible HIV-1 reservoir in U.S. patients compared to Ugandan individuals (48 vs. 11 cell equivalents/million cells, p < 0.0001). This divergence in reservoir size was verified measuring proviral DNA (206 vs. 88 cell equivalents/million cells, p < 0.0001). However, the peripheral HIV-1 reservoir was more diverse in Ugandan than in U.S. individuals (8.6 vs. 4.7 p-distance, p < 0.0001).
Conclusions: The smaller, but more diverse, peripheral HIV-1 reservoir in Ugandan patients might be associated with viral (e.g., non-B subtype with higher cytopathicity) and/or host (e.g., higher incidence of co-infections or co-morbidities leading to less clonal expansion) factors. This highlights the need to understand reservoir dynamics in diverse populations as part of ongoing efforts to find a functional cure for HIV-1 infection in LMICs.
Keywords: Co-infection; HIV; Reservoir; Subtype; Uganda.
© 2022. The Author(s).
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Paredes R, Clotet B. Clinical management of HIV-1 resistance. Antiviral Res. 2010;85(1):245–265. - PubMed
-
- Zolopa AR. The evolution of HIV treatment guidelines: current state-of-the-art of ART. Antiviral Res. 2010;85(1):241–244. - PubMed
-
- Saksena NK, Potter SJ. Reservoirs of HIV-1 in vivo: implications for antiretroviral therapy. AIDS Rev. 2003;5(1):3–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

