TLR4 downregulation by the RNA-binding protein PUM1 alleviates cellular aging and osteoarthritis
- PMID: 35034101
- PMCID: PMC9287402
- DOI: 10.1038/s41418-021-00925-6
TLR4 downregulation by the RNA-binding protein PUM1 alleviates cellular aging and osteoarthritis
Erratum in
-
Correction: TLR4 downregulation by the RNA-binding protein PUM1 alleviates cellular aging and osteoarthritis.Cell Death Differ. 2022 May;29(5):1077. doi: 10.1038/s41418-022-00945-w. Cell Death Differ. 2022. PMID: 35190709 Free PMC article. No abstract available.
Abstract
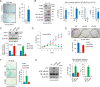
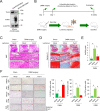
Dysfunction of mRNA or RNA-binding proteins (RBPs) causes cellular aging and age-related degenerative diseases; however, information regarding the mechanism through which RBP-mediated posttranscriptional regulation affects cellular aging and related disease processes is limited. In this study, PUM1 was found to be associated with the self-renewal capacity and aging process of human mesenchymal stem cells (MSC). PUM1 interacted with the 3'-untranslated region of Toll-like receptor 4 (TLR4) to suppress TLR4 mRNA translation and regulate the activity of nuclear factor-κB (NF-κB), a master regulator of the aging process in MSCs. PUM1 overexpression protected MSCs against H2O2-induced cellular senescence by suppressing TLR4-mediated NF-κB activity. TLR4-mediated NF-κB activation is a key regulator in osteoarthritis (OA) pathogenesis. PUM1 overexpression enhanced the chondrogenic potential of MSCs even under the influence of inflammation-inducing factors, such as lipopolysaccharide (LPS) or interleukin-1β (IL-1β), whereas the chondrogenic potential was reduced following the PUM1 knockdown-mediated TLR4 activation. PUM1 levels decreased under inflammatory conditions in vitro and during OA progression in human and mouse disease models. PUM1 knockdown in human chondrocytes promoted chondrogenic phenotype loss, whereas PUM1 overexpression protected the cells from inflammation-mediated disruption of the chondrogenic phenotype. Gene therapy using a lentiviral vector encoding mouse PUM1 showed promise in preserving articular cartilage integrity in OA mouse models. In conclusion, PUM1 is a novel suppressor of MSC aging, and the PUM1-TLR4 regulatory axis represents a potential therapeutic target for OA.
© 2021. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

