New Species and Eight Undescribed Species Belonging to the Families Aspergillaceae and Trichocomaceae in Korea
- PMID: 35035246
- PMCID: PMC8725871
- DOI: 10.1080/12298093.2021.1997461
New Species and Eight Undescribed Species Belonging to the Families Aspergillaceae and Trichocomaceae in Korea
Abstract
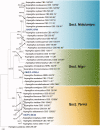
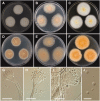
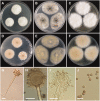
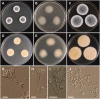
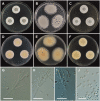
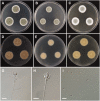
During a survey of fungal diversity associated with insects, mud, soil, and freshwater niches in different areas in Korea, nine interesting fungal strains were isolated. Based on their morphological characteristics and molecular phylogeny analyses, using a combined data set of β-tubulin (BenA), calmodulin (CaM), and second largest subunit of RNA polymerase (RPB2) sequences, the strains CNUFC AM-44, CNUFC JCW3-4, CNUFC S708, CNUFC WT202, CNUFC AS1-29, CNUFC JCW3-5, CNUFC JDP37, and CNUFC JDP62 were identified as Aspergillus alabamensis, A. floridensis, A. subversicolor, Penicillium flavigenum, P. laevigatum, P. lenticrescens, Talaromyces adpressus, and T. beijingensis, respectively. The strain CNUFC JT1301 belongs to series Westlingiorum in section Citrina and is phylogenetically related to P. manginii. However, slow growth when cultivated on CYA, MEA, CREA is observed and the property can be used to easily distinguish the new species from these species. Additionally, P. manginii is known to produce sclerotia, while CNUFC JT1301 strain does not. Herein, the new fungal species is proposed as P. aquadulcis sp. nov. Eight species, A. alabamensis, A. floridensis, A. subversicolor, P. flavigenum, P. laevigatum, P. lenticrescens, T. adpressus, and T. beijingensis, have not been previously reported in Korea. The present study expands the known distribution of fungal species belonging to the families Aspergillaceae and Trichocomaceae in Korea.
Keywords: Freshwater; insect; morphology; mud; phylogeny; soil; taxonomy.
© 2021 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group on behalf of the Korean Society of Mycology.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures







References
-
- Frisvad JC, Smedsgaard J, Larsen TO, et al. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004;49:201–241.
LinkOut - more resources
Full Text Sources
