SARS-CoV-2 Omicron Mutation Is Faster than the Chase: Multiple Mutations on Spike/ACE2 Interaction Residues
- PMID: 35036025
- PMCID: PMC8733186
- DOI: 10.4110/in.2021.21.e38
SARS-CoV-2 Omicron Mutation Is Faster than the Chase: Multiple Mutations on Spike/ACE2 Interaction Residues
Abstract
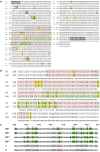
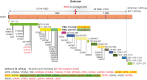
Recently, a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (B.1.1.529) Omicron variant originated from South Africa in the middle of November 2021. SARS-CoV-2 is also called coronavirus disease 2019 (COVID-19) since SARS-CoV-2 is the causative agent of COVID-19. Several studies already suggested that the SARS-CoV-2 Omicron variant would be the fastest transmissible variant compared to the previous 10 SARS-CoV-2 variants of concern, interest, and alert. Few clinical studies reported the high transmissibility of the Omicron variant but there is insufficient time to perform actual experiments to prove it, since the spread is so fast. We analyzed the SARS-CoV-2 Omicron variant, which revealed a very high rate of mutation at amino acid residues that interact with angiostatin-converting enzyme 2. The mutation rate of COVID-19 is faster than what we prepared vaccine program, antibody therapy, lockdown, and quarantine against COVID-19 so far. Thus, it is necessary to find better strategies to overcome the current crisis of COVID-19 pandemic.
Keywords: COVID-19 Omicron; Mutation; Receptor binding motif (RBM); SARS-CoV-2; Spike (S) gene.
Copyright © 2021. The Korean Association of Immunologists.
Conflict of interest statement
Conflict of Interest: The authors declare no potential conflicts of interest.
Figures
Similar articles
-
Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein.J Med Virol. 2022 Apr;94(4):1641-1649. doi: 10.1002/jmv.27526. Epub 2021 Dec 27. J Med Virol. 2022. PMID: 34914115
-
Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review.Eur Rev Med Pharmacol Sci. 2021 Dec;25(24):8019-8022. doi: 10.26355/eurrev_202112_27653. Eur Rev Med Pharmacol Sci. 2021. PMID: 34982466 Review.
-
The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem.J Biol Regul Homeost Agents. 2021 Jan-Feb;35(1):1-4. doi: 10.23812/21-3-E. J Biol Regul Homeost Agents. 2021. PMID: 33377359
-
Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance.World J Clin Cases. 2022 Jan 7;10(1):1-11. doi: 10.12998/wjcc.v10.i1.1. World J Clin Cases. 2022. PMID: 35071500 Free PMC article. Review.
-
SARS-CoV-2 B.1.1.529 (Omicron) Variant - United States, December 1-8, 2021.MMWR Morb Mortal Wkly Rep. 2021 Dec 17;70(50):1731-1734. doi: 10.15585/mmwr.mm7050e1. MMWR Morb Mortal Wkly Rep. 2021. PMID: 34914670 Free PMC article.
Cited by
-
Update on SARS-CoV-2 Omicron Variant of Concern and Its Peculiar Mutational Profile.Microbiol Spectr. 2022 Apr 27;10(2):e0273221. doi: 10.1128/spectrum.02732-21. Epub 2022 Mar 30. Microbiol Spectr. 2022. PMID: 35352942 Free PMC article.
-
First detection of SARS-CoV-2 Omicron BA.4 variant in Western Pennsylvania, United States.J Med Virol. 2022 Sep;94(9):4053-4055. doi: 10.1002/jmv.27846. Epub 2022 May 16. J Med Virol. 2022. PMID: 35534788 Free PMC article. No abstract available.
-
Pharmacoinformatic approach to identify potential phytochemicals against SARS-CoV-2 spike receptor-binding domain in native and variants of concern.Mol Divers. 2023 Dec;27(6):2741-2766. doi: 10.1007/s11030-022-10580-9. Epub 2022 Dec 22. Mol Divers. 2023. PMID: 36547813 Free PMC article.
-
Substitution spectra of SARS-CoV-2 genome from Pakistan reveals insights into the evolution of variants across the pandemic.Sci Rep. 2023 Nov 28;13(1):20955. doi: 10.1038/s41598-023-48272-5. Sci Rep. 2023. PMID: 38017265 Free PMC article.
-
Structural proteins of human coronaviruses: what makes them different?Front Cell Infect Microbiol. 2024 Dec 6;14:1458383. doi: 10.3389/fcimb.2024.1458383. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39711780 Free PMC article. Review.
References
-
- Brown CM, Vostok J, Johnson H, Burns M, Gharpure R, Sami S, Sabo RT, Hall N, Foreman A, Schubert PL, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings - Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1059–1062. - PMC - PubMed
-
- Dagpunar J. Interim estimates of increased transmissibility, growth rate, and reproduction number of the COVID-19 B.1.617.2 variant of concern in the United Kingdom. MedRxiv. 2021 doi: 10.1101/2021.06.03.21258293. - DOI
-
- Shi Q, Dong XP. Rapid global spread of the SARS-CoV-2 delta (B.1.617.2) variant: spatiotemporal variation and public health impact. Zoonoses. 2021;1:1–6.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

