Gene knock-out chain reaction enables high disruption efficiency of HPV18 E6/ E7 genes in cervical cancer cells
- PMID: 35036522
- PMCID: PMC8733033
- DOI: 10.1016/j.omto.2021.12.011
Gene knock-out chain reaction enables high disruption efficiency of HPV18 E6/ E7 genes in cervical cancer cells
Abstract
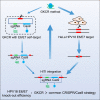
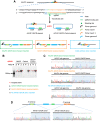
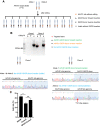
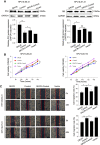
A genome editing tool targeting the high-risk human papillomavirus (HPV) oncogene is a promising therapeutic strategy to treat HPV-related cervical cancer. To improve gene knockout efficiency, we developed a gene knockout chain reaction (GKCR) method for continually generating mutagenic disruptions and used this method to disrupt the HPV18 E6 and E7 genes. We verified that the GKCR Cas9/guide RNA (gRNA) cassettes could integrated into the targeted loci via homology-independent targeted insertion (HITI). The qPCR results revealed that the GKCR method enabled a relatively higher Cas9/gRNA cassette insertion rate than a control method (the common CRISPR-Cas9 strategy). Tracking of Indels by DEcomposition (TIDE) assay results showed that the GKCR method produced a significantly higher percentage of insertions or deletions (indels) in the HPV18 E6 and E7 genes. Furthermore, by targeting the HPV18 E6/E7 oncogenes, we found that the GKCR method significantly upregulated the P53/RB proteins and inhibited the proliferation and motility of HeLa cells. The GKCR method significantly improved the gene knockout efficiency of the HPV18 E6/E7 oncogenes, which might provide new insights into treatment of HPV infection and related cervical cancer.
Keywords: CRISPR-Cas9; HITI; HPV; cervical cancer; gene knockout chain reaction (GKCR).
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. - PubMed
-
- Hu Z., Zhu D., Wang W., Li W., Jia W., Zeng X., Ding W., Yu L., Wang X., Wang L., et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015;47:158–163. - PubMed
-
- Hoppe-Seyler K., Bossler F., Braun J.A., Herrmann A.L., Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26:158–168. - PubMed
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

