Circadian REV-ERBs repress E4bp4 to activate NAMPT-dependent NAD+ biosynthesis and sustain cardiac function
- PMID: 35036997
- PMCID: PMC8754391
- DOI: 10.1038/s44161-021-00001-9
Circadian REV-ERBs repress E4bp4 to activate NAMPT-dependent NAD+ biosynthesis and sustain cardiac function
Abstract
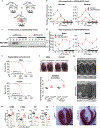
The heart is a highly metabolic organ that uses multiple energy sources to meet its demand for ATP production. Diurnal feeding-fasting cycles result in substrate availability fluctuations which, together with increased energetic demand during the active period, impose a need for rhythmic cardiac metabolism. The nuclear receptors REV-ERBα and β are essential repressive components of the molecular circadian clock and major regulators of metabolism. To investigate their role in the heart, here we generated mice with cardiomyocyte (CM)-specific deletion of both Rev-erbs, which died prematurely due to dilated cardiomyopathy. Loss of Rev-erbs markedly downregulated fatty acid oxidation genes prior to overt pathology, which was mediated by induction of the transcriptional repressor E4BP4, a direct target of cardiac REV-ERBs. E4BP4 directly controls circadian expression of Nampt and its biosynthetic product NAD+ via distal cis-regulatory elements. Thus, REV-ERB-mediated E4BP4 repression is required for Nampt expression and NAD+ production by the salvage pathway. Together, these results highlight the indispensable role of circadian REV-ERBs in cardiac gene expression, metabolic homeostasis and function.
Figures



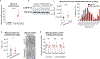







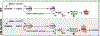
References
-
- Dierickx P et al. in Stem Cells and Cardiac Regeneration 57–78 (Springer International Publishing, 2015). doi:10.1007/978-3-319-25427-2_5 - DOI
-
- Crnko S, Pré, Du BC, Sluijter JPG & van Laake LW Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat Rev Cardiol 16, 437–447 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL128349/HL/NHLBI NIH HHS/United States
- R01 DK045586/DK/NIDDK NIH HHS/United States
- R01 DK098656/DK/NIDDK NIH HHS/United States
- R01 DK111495/DK/NIDDK NIH HHS/United States
- R01 HL143431/HL/NHLBI NIH HHS/United States
- R01 HL107594/HL/NHLBI NIH HHS/United States
- R01 HL149159/HL/NHLBI NIH HHS/United States
- F32 DK122684/DK/NIDDK NIH HHS/United States
- R01 HL151345/HL/NHLBI NIH HHS/United States
- R01 HL165792/HL/NHLBI NIH HHS/United States
- P30 DK019525/DK/NIDDK NIH HHS/United States
- F32 HL145923/HL/NHLBI NIH HHS/United States
- R01 HL058493/HL/NHLBI NIH HHS/United States
- P30 DK079626/DK/NIDDK NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
