Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments
- PMID: 35038580
- PMCID: PMC8899700
- DOI: 10.1016/j.ymthe.2022.01.022
Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments
Abstract
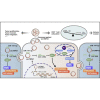
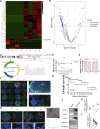
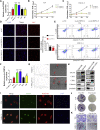
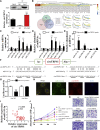
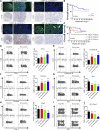
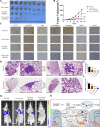
Circular RNAs (circRNAs) play critical roles in different diseases. Exosomes are important intermediates of intercellular communication. While both have been widely reported in cancers, exosome-derived circRNAs are rarely studied. In this work, we identified the differently expressed circRNAs in bladder cancer (BCa) tissue and exosomes through high-throughput sequencing. RNA pull-down, RNA immunoprecipitation, and luciferase reporter assays were used to investigate the interactions between specific circRNAs, microRNAs (miRNAs), and mRNAs. Wound-healing, Transwell, Cell Counting Kit-8 (CCK8), and colony-formation assays were used to study the biological roles in vitro. Metabolomics were used to explore the mechanism of how specific circRNAs influenced BCa cell behavior. Flow cytometry was used to study how specific circRNAs affected the function of CD8+ T cells in tumor microenvironments. We identified that exosome-derived hsa_circ_0085361 (circTRPS1) was correlated with aggressive phenotypes of BCa cells via sponging miR-141-3p. Metabolomics and RNA sequencing (RNA-seq) identified GLS1-mediated glutamine metabolism was involved in circTRPS1-mediated alterations. Exosomes derived from circTRPS1 knocked down BCa cells, prevented CD8+ T cells from exhaustion, and repressed the malignant phenotype of BCa cells. In conclusion, exosome-derived circTRPS1 from BCa cells can modulate the intracellular reactive oxygen species (ROS) balance and CD8+ T cell exhaustion via the circTRPS1/miR141-3p/GLS1 axis. Our work may provide a potential biomarker and therapeutic target for BCa.
Keywords: CD8+ T cell; ROS; bladder cancer; circular RNAs; exosomes; tumor microenvironment.
Copyright © 2022 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. - PubMed
-
- Nekolla K., Brieu N., Gavriel C.G., Widmaier M., Budco A., Medrikova D., Kanchev I., Testori M., Chan J., Dundee P., et al. Prognostic immunoprofiling of muscle invasive bladder cancer (MIBC) patients in a multicentre setting. Ann. Oncol. 2019;30:v49–v50.
-
- Knowles M.A., Hurst C.D. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat. Rev. Cancer. 2015;15(1):25–41. - PubMed
-
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9(6):654–659. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

