A Filamentous Bacteriophage Protein Inhibits Type IV Pili To Prevent Superinfection of Pseudomonas aeruginosa
- PMID: 35038902
- PMCID: PMC8764522
- DOI: 10.1128/mbio.02441-21
A Filamentous Bacteriophage Protein Inhibits Type IV Pili To Prevent Superinfection of Pseudomonas aeruginosa
Abstract
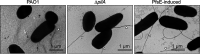
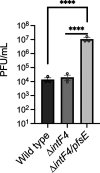
Pseudomonas aeruginosa is an opportunistic pathogen that causes infections in a variety of settings. Many P. aeruginosa isolates are infected by filamentous Pf bacteriophage integrated into the bacterial chromosome as a prophage. Pf virions can be produced without lysing P. aeruginosa. However, cell lysis can occur during superinfection, which occurs when Pf virions successfully infect a host lysogenized by a Pf prophage. Temperate phages typically encode superinfection exclusion mechanisms to prevent host lysis by virions of the same or similar species. In this study, we sought to elucidate the superinfection exclusion mechanism of Pf phage. Initially, we observed that P. aeruginosa that survive Pf superinfection are transiently resistant to Pf-induced plaquing and are deficient in twitching motility, which is mediated by type IV pili (T4P). Pf utilize T4P as a cell surface receptor, suggesting that T4P are suppressed in bacteria that survive superinfection. We tested the hypothesis that a Pf-encoded protein suppresses T4P to mediate superinfection exclusion by expressing Pf proteins in P. aeruginosa and measuring plaquing and twitching motility. We found that the Pf protein PA0721, which we termed
Keywords: Pf4; PilC; Pseudomonas aeruginosa; filamentous bacteriophage; superinfection exclusion; twitching motility; type IV pili.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Secor PR, Burgener EB, Kinnersley M, Jennings LK, Roman-Cruz V, Popescu M, Van Belleghem JD, Haddock N, Copeland C, Michaels LA, de Vries CR, Chen Q, Pourtois J, Wheeler TJ, Milla CE, Bollyky PL. 2020. Pf bacteriophage and their impact on pseudomonas virulence, mammalian immunity, and chronic infections. Front Immunol 11:244. doi:10.3389/fimmu.2020.00244. - DOI - PMC - PubMed
-
- Burgener EB, Sweere JM, Bach MS, Secor PR, Haddock N, Jennings LK, Marvig RL, Johansen HK, Rossi E, Cao X, Tian L, Nedelec L, Molin S, Bollyky PL, Milla CE. 2019. Filamentous bacteriophages are associated with chronic Pseudomonas lung infections and antibiotic resistance in cystic fibrosis. Sci Transl Med 11:eaau9748. doi:10.1126/scitranslmed.aau9748. - DOI - PMC - PubMed
-
- Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, Sunkari V, Kaber G, Manasherob R, Suh GA, Cao X, de Vries CR, Lam DN, Marshall PL, Birukova M, Katznelson E, Lazzareschi DV, Balaji S, Keswani SG, Hawn TR, Secor PR, Bollyky PL. 2019. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363:6513. doi:10.1126/science.aat9691. - DOI - PMC - PubMed
-
- Secor PR, Michaels LA, Smigiel KS, Rohani MG, Jennings LK, Hisert KB, Arrigoni A, Braun KR, Birkland TP, Lai Y, Hallstrand TS, Bollyky PL, Singh PK, Parks WC. 2017. Filamentous bacteriophage produced by Pseudomonas aeruginosa alters the inflammatory response and promotes noninvasive infection in vivo. Infect Immun 85:e00648-16. doi:10.1128/IAI.00648-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
