Genetic Dissection of Antibiotic Adjuvant Activity
- PMID: 35038910
- PMCID: PMC8764523
- DOI: 10.1128/mbio.03084-21
Genetic Dissection of Antibiotic Adjuvant Activity
Abstract
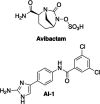
Small molecule adjuvants that enhance the activity of established antibiotics represent promising agents in the battle against antibiotic resistance. Adjuvants generally act by inhibiting antibiotic resistance processes, and specifying the process acted on is a critical step in defining an adjuvant's mechanism of action. This step is typically carried out biochemically by identifying molecules that bind adjuvants and then inferring their roles in resistance. Here, we present a complementary genetic strategy based on identifying mutations that both sensitize cells to antibiotic and make them "adjuvant blind." We tested the approach in Acinetobacter baumannii AB5075 using two adjuvants: a well-characterized β-lactamase inhibitor (avibactam) and a compound enhancing outer membrane permeability (aryl 2-aminoimidazole AI-1). The avibactam studies showed that the adjuvant potentiated one β-lactam (ceftazidime) through action on a single β-lactamase (GES-14) and a second (meropenem) by targeting two different enzymes (GES-14 and OXA-23). Mutations impairing disulfide bond formation (DsbAB) also reduced potentiation, possibly by impairing β-lactamase folding. Mutations reducing AI-1 potentiation of canonical Gram-positive antibiotics (vancomycin and clarithromycin) blocked lipooligosaccharide (LOS/LPS) synthesis or its acyl modification. The results indicate that LOS-mediated outer membrane impermeability is targeted by the adjuvant and show the importance of acylation in the resistance. As part of the study, we employed Acinetobacter baylyi as a model to verify the generality of the A. baumannii results and identified the principal resistance genes for ceftazidime, meropenem, vancomycin, and clarithromycin in A. baumannii AB5075. Overall, the work provides a foundation for analyzing adjuvant action using a comprehensive genetic approach. IMPORTANCE One strategy to confront the antibiotic resistance crisis is through the development of adjuvant compounds that increase the efficacy of established drugs. A key step in the development of a natural product adjuvant as a drug is identifying the resistance process it undermines to enhance antibiotic activity. Previous procedures designed to accomplish this have relied on biochemical identification of cell components that bind adjuvant. Here, we present a complementary strategy based on identifying mutations that eliminate adjuvant activity.
Keywords: Acinetobacter; Tn-seq; aminoimidazole; avibactam; baumannii; baylyi; meropenem; vancomycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

