ClinGen Variant Curation Interface: a variant classification platform for the application of evidence criteria from ACMG/AMP guidelines
- PMID: 35039090
- PMCID: PMC8764818
- DOI: 10.1186/s13073-021-01004-8
ClinGen Variant Curation Interface: a variant classification platform for the application of evidence criteria from ACMG/AMP guidelines
Abstract
Background: Identification of clinically significant genetic alterations involved in human disease has been dramatically accelerated by developments in next-generation sequencing technologies. However, the infrastructure and accessible comprehensive curation tools necessary for analyzing an individual patient genome and interpreting genetic variants to inform healthcare management have been lacking.
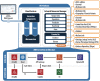
Results: Here we present the ClinGen Variant Curation Interface (VCI), a global open-source variant classification platform for supporting the application of evidence criteria and classification of variants based on the ACMG/AMP variant classification guidelines. The VCI is among a suite of tools developed by the NIH-funded Clinical Genome Resource (ClinGen) Consortium and supports an FDA-recognized human variant curation process. Essential to this is the ability to enable collaboration and peer review across ClinGen Expert Panels supporting users in comprehensively identifying, annotating, and sharing relevant evidence while making variant pathogenicity assertions. To facilitate evidence-based improvements in human variant classification, the VCI is publicly available to the genomics community. Navigation workflows support users providing guidance to comprehensively apply the ACMG/AMP evidence criteria and document provenance for asserting variant classifications.
Conclusions: The VCI offers a central platform for clinical variant classification that fills a gap in the learning healthcare system, facilitates widespread adoption of standards for clinical curation, and is available at https://curation.clinicalgenome.org.
Keywords: Clinical Genome Resource Consortium; Clinical genetics; Precision medicine; Variant curation.
© 2021. The Author(s).
Conflict of interest statement
S.E.P. is a member of the Baylor Genetics Scientific Advisory Panel. A.M. is an employee of Baylor College of Medicine (BCM) and performs integration consulting services for BCM-developed software including Genboree through IP Genesis Inc. C.D.B. is on the scientific advisory boards (SAB) of AncestryDNA, Arc Bio LLC, Etalon DX, Liberty Biosecurity, and Personalis. C.D.B. is on the board of EdenRoc Sciences LLC. C.D.B. is also a founder and SAB chair of ARCBio. J.L.M. is an employee of GeneDx/BioReference Laboratories, Inc./OPKO Health and has a salary as the only disclosure. None of these entities played a role in the design, execution, interpretation, or presentation of this study. The remaining authors declare that they have no competing interests.
Figures



References
-
- Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

