Malaria abrogates O'nyong-nyong virus pathologies by restricting virus infection in nonimmune cells
- PMID: 35039441
- PMCID: PMC8807878
- DOI: 10.26508/lsa.202101272
Malaria abrogates O'nyong-nyong virus pathologies by restricting virus infection in nonimmune cells
Abstract
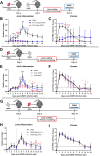
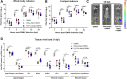
O'nyongnyong virus (ONNV) is a re-emerging alphavirus previously known to be transmitted by main malaria vectors, thus suggesting the possibility of coinfections with arboviruses in co-endemic areas. However, the pathological outcomes of such infections remain unknown. Using murine coinfection models, we demonstrated that a preexisting blood-stage Plasmodium infection suppresses ONNV-induced pathologies. We further showed that suppression of viremia and virus dissemination are dependent on Plasmodium-induced IFNγ and are associated with reduced infection of CD45- cells at the site of virus inoculation. We further proved that treatment with IFNγ or plasma samples from Plasmodium vivax-infected patients containing IFNγ are able to restrict ONNV infection in human fibroblast, synoviocyte, skeletal muscle, and endothelial cell lines. Mechanistically, the role of IFNγ in restricting ONNV infection was confirmed in in vitro infection assays through the generation of an IFNγ receptor 1 α chain (IFNγR1)-deficient cell line.
© 2022 Torres-Ruesta et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures










References
-
- Sanders EJ, Rwaguma EB, Kawamata J, Kiwanuka N, Lutwama JJ, Ssengooba FP, Lamunu M, Najjemba R, Were WA, Bagambisa G, et al. (1999) O’nyong-nyong fever in south-central Uganda, 1996-1997: Description of the epidemic and results of a household-based seroprevalence survey. J Infect Dis 180: 1436–1443. 10.1086/315073 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
