Heterogeneous digital biomarker integration out-performs patient self-reports in predicting Parkinson's disease
- PMID: 35039601
- PMCID: PMC8763910
- DOI: 10.1038/s42003-022-03002-x
Heterogeneous digital biomarker integration out-performs patient self-reports in predicting Parkinson's disease
Abstract
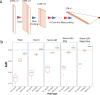
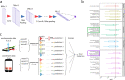
Parkinson's disease (PD) is one of the first diseases where digital biomarkers demonstrated excellent performance in differentiating disease from healthy individuals. However, no study has systematically compared and leveraged multiple types of digital biomarkers to predict PD. Particularly, machine learning works on the fine-motor skills of PD are limited. Here, we developed deep learning methods that achieved an AUC (Area Under the receiver operator characteristic Curve) of 0.933 in identifying PD patients on 6418 individuals using 75048 tapping accelerometer and position records. Performance of tapping is superior to gait/rest and voice-based models obtained from the same benchmark population. Assembling the three models achieved a higher AUC of 0.944. Notably, the models not only correlated strongly to, but also performed better than patient self-reported symptom scores in diagnosing PD. This study demonstrates the complementary predictive power of tapping, gait/rest and voice data and establishes integrative deep learning-based models for identifying PD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: J.W. is a current Eli Lilly and Company employee. Y.G. serves as the scientific advisor for Eli Lilly and Company on the tapping part of this study. R.L.A. serves on the DSMBs for the COMPASS and PASSPORT trials (Biogen), the M-STAR trial (Biohaven), and the Signal-AD trial (Vaccinex). R.L.A. has received consulting fees from the Michael J. Fox Foundation and Takeda. The remaining authors declare no competing interests.
Figures




References
-
- Frank, A. & Asuncion, A. UCI Machine Learning Repository (University of California, 2010).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

