Nuclear and peroxisomal targeting of catalase
- PMID: 35040158
- PMCID: PMC9305541
- DOI: 10.1111/pce.14262
Nuclear and peroxisomal targeting of catalase
Abstract
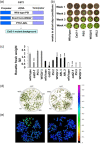
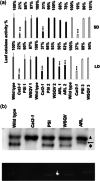
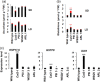
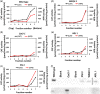
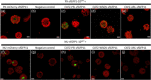
Catalase is a well-known component of the cellular antioxidant network, but there have been conflicting conclusions reached regarding the nature of its peroxisome targeting signal. It has also been reported that catalase can be hijacked to the nucleus by effector proteins of plant pathogens. Using a physiologically relevant system where native untagged catalase variants are expressed in a cat2-1 mutant background, the C terminal most 18 amino acids could be deleted without affecting activity, peroxisomal targeting or ability to complement multiple phenotypes of the cat2-1 mutant. In contrast, converting the native C terminal tripeptide PSI to the canonical PTS1 sequence ARL resulted in lower catalase specific activity. Localisation experiments using split superfolder green fluorescent protein revealed that catalase can be targeted to the nucleus in the absence of any pathogen effectors, and that C terminal tagging in combination with alterations of the native C terminus can interfere with nuclear localisation. These findings provide fundamental new insights into catalase targeting and pave the way for exploration of the mechanism of catalase targeting to the nucleus and its role in non-infected plants.
Keywords: ROS; nucleus; peroxisome; redox signalling.
© 2022 The Authors. Plant, Cell & Environment published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures

References
-
- Brocard, C. & Hartig, A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochimica et Biophysica Acta (BBA)‐Molecular Cell Research, 1763(12), 1565–1573. - PubMed
-
- Clough, S.J. & Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16(6), 735–743. - PubMed
-
- Dietz, K.‐J. , Turkan, I. & Krieger‐Liszkay, A. (2016) Redox‐and reactive oxygen species‐dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiology, 171(3), 1541–1550. https://academic.oup.com/plphys/article/171/3/1541/6115116 - PMC - PubMed
-
- Freitas, M.O. , Francisco, T. , Rodrigues, T.A. , Alencastre, I.S. , Pinto, M.P. , Grou, C.P. et al. (2011) PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N‐terminal domain of PEX14. Journal of Biological Chemistry, 286(47), 40509–40519. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

