Genetic basis and dual adaptive role of floral pigmentation in sunflowers
- PMID: 35040432
- PMCID: PMC8765750
- DOI: 10.7554/eLife.72072
Genetic basis and dual adaptive role of floral pigmentation in sunflowers
Abstract
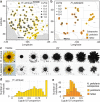
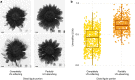
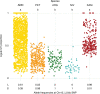
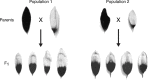
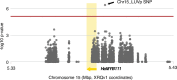
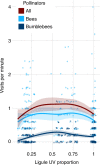
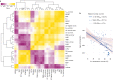
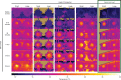
Variation in floral displays, both between and within species, has been long known to be shaped by the mutualistic interactions that plants establish with their pollinators. However, increasing evidence suggests that abiotic selection pressures influence floral diversity as well. Here, we analyse the genetic and environmental factors that underlie patterns of floral pigmentation in wild sunflowers. While sunflower inflorescences appear invariably yellow to the human eye, they display extreme diversity for patterns of ultraviolet pigmentation, which are visible to most pollinators. We show that this diversity is largely controlled by cis-regulatory variation affecting a single MYB transcription factor, HaMYB111, through accumulation of ultraviolet (UV)-absorbing flavonol glycosides in ligules (the 'petals' of sunflower inflorescences). Different patterns of ultraviolet pigments in flowers are strongly correlated with pollinator preferences. Furthermore, variation for floral ultraviolet patterns is associated with environmental variables, especially relative humidity, across populations of wild sunflowers. Ligules with larger ultraviolet patterns, which are found in drier environments, show increased resistance to desiccation, suggesting a role in reducing water loss. The dual role of floral UV patterns in pollinator attraction and abiotic response reveals the complex adaptive balance underlying the evolution of floral traits.
Keywords: abiotic stress; adaptation; evolutionary biology; floral pigmentation; genetics; genomics; pollination; sunflower; sunflower (helianthus annuus); transcription factors.
Plain language summary
Flowers are an important part of how many plants reproduce. Their distinctive colours, shapes and patterns attract specific pollinators, but they can also help to protect the plant from predators and environmental stresses. Many flowers contain pigments that absorb ultraviolet (UV) light to display distinct UV patterns – although invisible to the human eye, most pollinators are able to see them. For example, when seen in UV, sunflowers feature a ‘bullseye’ with a dark centre surrounded by a reflective outer ring. The sizes and thicknesses of these rings vary a lot within and between flower species, and so far, it has been unclear what causes this variation and how it affects the plants. To find out more, Todesco et al. studied the UV patterns in various wild sunflowers across North America by considering the ecology and molecular biology of different plants. This revealed great variation between the UV patterns of the different sunflower populations. Moreover, Todesco et al. found that a gene called HaMYB111 is responsible for the diverse UV patterns in the sunflowers. This gene controls how plants make chemicals called flavonols that absorb UV light. Flavonols also help to protect plants from damage caused by droughts and extreme temperatures. Todesco et al. showed that plants with larger bullseyes had more flavonols, attracted more pollinators, and were better at conserving water. Accordingly, these plants were found in drier locations. This study suggests that, at least in sunflowers, UV patterns help both to attract pollinators and to control water loss. These insights could help to improve pollination – and consequently yield – in cultivated plants, and to develop plants with better resistance to extreme weather. This work also highlights the importance of combining biology on small and large scales to understand complex processes, such as adaptation and evolution.
© 2022, Todesco et al.
Conflict of interest statement
MT, NB, AK, II, GO, ÓD, SH, LM, MJ, JL, BB, LR No competing interests declared
Figures








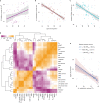
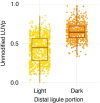
Comment in
-
Why do sunflowers have invisible colors?Elife. 2022 Jan 21;11:e76105. doi: 10.7554/eLife.76105. Elife. 2022. PMID: 35061584 Free PMC article.
References
-
- Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B, Legrand L, Gill N, Kane NC, Bowers JE, Hubner S, Bellec A, Bérard A, Bergès H, Blanchet N, Boniface M-C, Brunel D, Catrice O, Chaidir N, Claudel C, Donnadieu C, Faraut T, Fievet G, Helmstetter N, King M, Knapp SJ, Lai Z, Le Paslier M-C, Lippi Y, Lorenzon L, Mandel JR, Marage G, Marchand G, Marquand E, Bret-Mestries E, Morien E, Nambeesan S, Nguyen T, Pegot-Espagnet P, Pouilly N, Raftis F, Sallet E, Schiex T, Thomas J, Vandecasteele C, Varès D, Vear F, Vautrin S, Crespi M, Mangin B, Burke JM, Salse J, Muños S, Vincourt P, Rieseberg LH, Langlade NB. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. - DOI - PubMed
-
- Beckmann M, Václavík T, Manceur AM, Šprtová L, von Wehrden H, Welk E, Cord AF, Tatem A. glUV: a global UV-B radiation data set for macroecological studies. Methods in Ecology and Evolution. 2014;5:372–383. doi: 10.1111/2041-210X.12168. - DOI
-
- Bock DG, Kantar MB, Rieseberg L. Population GGenomics of SSpeciation and AAdaptation in SSunflowers. Springer; 2020. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

