Longitudinally monitored immune biomarkers predict the timing of COVID-19 outcomes
- PMID: 35041647
- PMCID: PMC8812869
- DOI: 10.1371/journal.pcbi.1009778
Longitudinally monitored immune biomarkers predict the timing of COVID-19 outcomes
Abstract
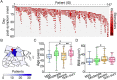
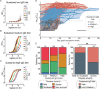
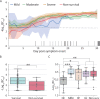
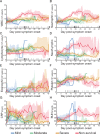
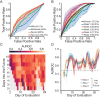
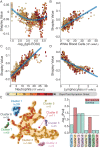
The clinical outcome of SARS-CoV-2 infection varies widely between individuals. Machine learning models can support decision making in healthcare by assessing fatality risk in patients that do not yet show severe signs of COVID-19. Most predictive models rely on static demographic features and clinical values obtained upon hospitalization. However, time-dependent biomarkers associated with COVID-19 severity, such as antibody titers, can substantially contribute to the development of more accurate outcome models. Here we show that models trained on immune biomarkers, longitudinally monitored throughout hospitalization, predicted mortality and were more accurate than models based on demographic and clinical data upon hospital admission. Our best-performing predictive models were based on the temporal analysis of anti-SARS-CoV-2 Spike IgG titers, white blood cell (WBC), neutrophil and lymphocyte counts. These biomarkers, together with C-reactive protein and blood urea nitrogen levels, were found to correlate with severity of disease and mortality in a time-dependent manner. Shapley additive explanations of our model revealed the higher predictive value of day post-symptom onset (PSO) as hospitalization progresses and showed how immune biomarkers contribute to predict mortality. In sum, we demonstrate that the kinetics of immune biomarkers can inform clinical models to serve as a powerful monitoring tool for predicting fatality risk in hospitalized COVID-19 patients, underscoring the importance of contextualizing clinical parameters according to their time post-symptom onset.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interets: K.C. is a member of the scientific advisory board of Integrum Scientific, LLC. J.R.L is a consultant for Celdara Medical.
Figures
Similar articles
-
Cytokine Profiles Associated With Worse Prognosis in a Hospitalized Peruvian COVID-19 Cohort.Front Immunol. 2021 Sep 1;12:700921. doi: 10.3389/fimmu.2021.700921. eCollection 2021. Front Immunol. 2021. PMID: 34539631 Free PMC article.
-
Clinical characteristics and antibody response to SARS-CoV-2 spike 1 protein using VITROS Anti-SARS-CoV-2 antibody tests in COVID-19 patients in Japan.J Med Microbiol. 2021 Apr;70(4):001291. doi: 10.1099/jmm.0.001291. J Med Microbiol. 2021. PMID: 33861191 Free PMC article.
-
Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: a retrospective study.Emerg Microbes Infect. 2021 Dec;10(1):664-676. doi: 10.1080/22221751.2021.1905488. Emerg Microbes Infect. 2021. PMID: 33734013 Free PMC article.
-
Interplay of Antibody and Cytokine Production Reveals CXCL13 as a Potential Novel Biomarker of Lethal SARS-CoV-2 Infection.mSphere. 2021 Jan 20;6(1):e01324-20. doi: 10.1128/mSphere.01324-20. mSphere. 2021. PMID: 33472985 Free PMC article.
-
Quantitative SARS-CoV-2 Serology in Children With Multisystem Inflammatory Syndrome (MIS-C).Pediatrics. 2020 Dec;146(6):e2020018242. doi: 10.1542/peds.2020-018242. Epub 2020 Sep 2. Pediatrics. 2020. PMID: 32879033
Cited by
-
Dynamical modelling of viral infection and cooperative immune protection in COVID-19 patients.PLoS Comput Biol. 2023 Sep 1;19(9):e1011383. doi: 10.1371/journal.pcbi.1011383. eCollection 2023 Sep. PLoS Comput Biol. 2023. PMID: 37656752 Free PMC article.
-
Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study.J Cardiovasc Dev Dis. 2023 Jan 23;10(2):39. doi: 10.3390/jcdd10020039. J Cardiovasc Dev Dis. 2023. PMID: 36826535 Free PMC article.
-
Comment to "Very late intubation in COVID-19 patients: A forgotten prognosis factor?".Crit Care. 2022 Jul 11;26(1):212. doi: 10.1186/s13054-022-04033-w. Crit Care. 2022. PMID: 35818055 Free PMC article. No abstract available.
-
Laboratory findings predictive of critical illness in hospitalized COVID-19 patients in Tunisia.F1000Res. 2024 Nov 18;13:918. doi: 10.12688/f1000research.151333.2. eCollection 2024. F1000Res. 2024. PMID: 39659435 Free PMC article.
-
SERS-Based Biosensors Combined with Machine Learning for Medical Application.ChemistryOpen. 2023 Jan;12(1):e202200192. doi: 10.1002/open.202200192. ChemistryOpen. 2023. PMID: 36627171 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

