Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa
- PMID: 35042229
- PMCID: PMC8942855
- DOI: 10.1038/s41586-022-04411-y
Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa
Abstract
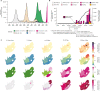
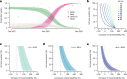
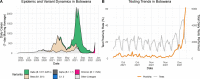
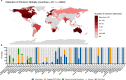
The SARS-CoV-2 epidemic in southern Africa has been characterized by three distinct waves. The first was associated with a mix of SARS-CoV-2 lineages, while the second and third waves were driven by the Beta (B.1.351) and Delta (B.1.617.2) variants, respectively1-3. In November 2021, genomic surveillance teams in South Africa and Botswana detected a new SARS-CoV-2 variant associated with a rapid resurgence of infections in Gauteng province, South Africa. Within three days of the first genome being uploaded, it was designated a variant of concern (Omicron, B.1.1.529) by the World Health Organization and, within three weeks, had been identified in 87 countries. The Omicron variant is exceptional for carrying over 30 mutations in the spike glycoprotein, which are predicted to influence antibody neutralization and spike function4. Here we describe the genomic profile and early transmission dynamics of Omicron, highlighting the rapid spread in regions with high levels of population immunity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
COVID-19 wave 4 in Western Cape Province, South Africa: Fewer hospitalisations, but new challenges for a depleted workforce.S Afr Med J. 2022 Feb 1;112(2):13496. S Afr Med J. 2022. PMID: 35139985 No abstract available.
References
-
- Tegally H, et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. - PubMed
-
- Tegally H, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. - PubMed
-
- Tegally, H. et al. Rapid replacement of the Beta variant by the Delta variant in South Africa. Preprint at medRxiv10.1101/2021.09.23.21264018 (2021).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- ERC_/European Research Council/International
- K24 AI131928/AI/NIAID NIH HHS/United States
- U01IP001048/ACL/ACL HHS/United States
- 221003/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- D43 TW009610/TW/FIC NIH HHS/United States
- 222574/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- U01 AI151698/AI/NIAID NIH HHS/United States
- U01 IP001048/IP/NCIRD CDC HHS/United States
- 203135/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 222754/WT_/Wellcome Trust/United Kingdom
- K24 AI131924/AI/NIAID NIH HHS/United States
- 206298/Z/17/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

