Daughter centrioles assemble preferentially towards the nuclear envelope in Drosophila syncytial embryos
- PMID: 35042404
- PMCID: PMC8767211
- DOI: 10.1098/rsob.210343
Daughter centrioles assemble preferentially towards the nuclear envelope in Drosophila syncytial embryos
Abstract
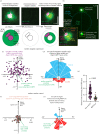
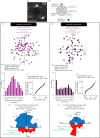
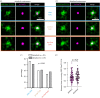
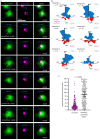
Centrosomes are important organizers of microtubules within animal cells. They comprise a pair of centrioles surrounded by the pericentriolar material, which nucleates and organizes the microtubules. To maintain centrosome numbers, centrioles must duplicate once and only once per cell cycle. During S-phase, a single new 'daughter' centriole is built orthogonally on one side of each radially symmetric 'mother' centriole. Mis-regulation of duplication can result in the simultaneous formation of multiple daughter centrioles around a single mother centriole, leading to centrosome amplification, a hallmark of cancer. It remains unclear how a single duplication site is established. It also remains unknown whether this site is pre-defined or randomly positioned around the mother centriole. Here, we show that within Drosophila syncytial embryos daughter centrioles preferentially assemble on the side of the mother facing the nuclear envelope, to which the centrosomes are closely attached. This positional preference is established early during duplication and remains stable throughout daughter centriole assembly, but is lost in centrosomes forced to lose their connection to the nuclear envelope. This shows that non-centrosomal cues influence centriole duplication and raises the possibility that these external cues could help establish a single duplication site.
Keywords: Drosophila; centriole; centriole duplication; centrosome; microtubules.
Conflict of interest statement
We declare we have no competing interests.
Figures
Similar articles
-
Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts.Curr Biol. 2010 Dec 21;20(24):2187-92. doi: 10.1016/j.cub.2010.11.055. Epub 2010 Dec 9. Curr Biol. 2010. PMID: 21145745
-
Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos.Curr Biol. 2014 Jun 2;24(11):1276-82. doi: 10.1016/j.cub.2014.04.023. Epub 2014 May 15. Curr Biol. 2014. PMID: 24835456 Free PMC article.
-
Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes.Dev Cell. 2016 Jun 20;37(6):545-57. doi: 10.1016/j.devcel.2016.05.022. Dev Cell. 2016. PMID: 27326932 Free PMC article.
-
Centriole inheritance.Prion. 2008 Jan-Mar;2(1):9-16. doi: 10.4161/pri.2.1.5064. Epub 2008 Jan 12. Prion. 2008. PMID: 19164929 Free PMC article. Review.
-
Accessorizing the centrosome: new insights into centriolar appendages and satellites.Curr Opin Struct Biol. 2021 Feb;66:148-155. doi: 10.1016/j.sbi.2020.10.021. Epub 2020 Dec 3. Curr Opin Struct Biol. 2021. PMID: 33279729 Review.
Cited by
-
Duplication and Segregation of Centrosomes during Cell Division.Cells. 2022 Aug 7;11(15):2445. doi: 10.3390/cells11152445. Cells. 2022. PMID: 35954289 Free PMC article. Review.
-
Centrosome Formation in the Bovine Early Embryo.Cells. 2023 May 7;12(9):1335. doi: 10.3390/cells12091335. Cells. 2023. PMID: 37174735 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

