Molecular characterization of Trypanosoma evansi, T. vivax and T. congolense in camels (Camelus dromedarius) of KSA
- PMID: 35042521
- PMCID: PMC8764778
- DOI: 10.1186/s12917-022-03148-0
Molecular characterization of Trypanosoma evansi, T. vivax and T. congolense in camels (Camelus dromedarius) of KSA
Abstract
Background: Trypanosoma evansi is the leading infectious Trypanosoma spp. in camels (Camelus dromedarius) present in the Kingdom of Saudi Arabia (KSA) that could lead to extensive economic losses. The present study was aimed to assess the prevalence rate of T. evansi in Taif governorate, Makkah province, KSA using parasitological and molecular evaluations, and analyze their genetic relationship targeting internal transcribed spacer 1 (ITS1) and variable surface glycoprotein (VSG) genes. For evaluation, we have used 102 blood samples of camels obtained from three different regions in Taif.
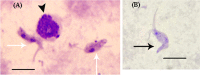
Results: Results show a considerable prevalence rate of trypanosomosis 2/102 (2.0%) according to Giemsa-stained buffy coat smear, and 16/102 (15.7%) according to touchdown PCR. T. evansi (n = 10/102, 9.8%) was the main infectious species found in camels then T. vivax (n = 3/102, 2.9%). Mixed infections were detected in three camels with T. evansi, T. vivax, and T. congolense (n = 3/102, 2.9%). Regarding gender, the results indicate that female camels (11/66, 16.7%) show higher prevalence of Trypanosoma than males (5/36, 13.9%). Sequencing and phylogenetic analyses of ITS1 and VSG showed their relationships with T. evansi in other hosts from different countries.
Conclusions: In our peer knowledge, it is the first time to report a research-based prevalence of trypanosomosis in the camels of Taif governorate, Makkah province, KSA.
Keywords: ITS1; KSA; Phylogeny; Rotat 1.2 VSG; Taif governorate; Trypanosomosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- FAO-statistics. Available from: http://faostat.fao.org. 2019.
-
- Gaili ESE, Al-Eknah M, Mansour H. Systems of camel management in Saudi Arabia. Arab J Agric Res. 2000;116:148–156.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

