The asymmetric distribution of RNA polymerase II and nucleosomes on replicated daughter genomes is caused by differences in replication timing between the lagging and the leading strand
- PMID: 35042724
- PMCID: PMC8805712
- DOI: 10.1101/gr.275387.121
The asymmetric distribution of RNA polymerase II and nucleosomes on replicated daughter genomes is caused by differences in replication timing between the lagging and the leading strand
Abstract
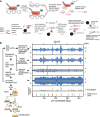
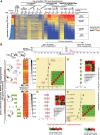
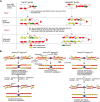
Chromatin features are thought to have a role in the epigenetic transmission of transcription states from one cell generation to the next. It is unclear how chromatin structure survives disruptions caused by genomic replication or whether chromatin features are instructive of the transcription state of the underlying gene. We developed a method to monitor budding yeast replication, transcription, and chromatin maturation dynamics on each daughter genome in parallel, with which we identified clusters of secondary origins surrounding known origins. We found a difference in the timing of lagging and leading strand replication on the order of minutes at most yeast genes. We propose a model in which the majority of old histones and RNA polymerase II (RNAPII) bind to the gene copy that replicated first, while newly synthesized nucleosomes are assembled on the copy that replicated second. RNAPII enrichment then shifts to the sister copy that replicated second. The order of replication is largely determined by genic orientation: If transcription and replication are codirectional, the leading strand replicates first; if they are counterdirectional, the lagging strand replicates first. A mutation in the Mcm2 subunit of the replicative helicase Mcm2-7 that impairs Mcm2 interactions with histone H3 slows down replication forks but does not qualitatively change the asymmetry in nucleosome distribution observed in the WT. We propose that active transcription states are inherited simultaneously and independently of their underlying chromatin states through the recycling of the transcription machinery and old histones, respectively. Transcription thus actively contributes to the reestablishment of the active chromatin state.
© 2022 Ziane et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Blank HM, Papoulas O, Maitra N, Garge R, Kennedy BK, Schilling B, Marcotte EM, Polymenis M. 2020. Abundances of transcripts, proteins, and metabolites in the cell cycle of budding yeast reveal coordinate control of lipid metabolism. Mol Biol Cell 31: 1069–1084. 10.1091/mbc.E19-12-0708 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
