Darwinian genomics and diversity in the tree of life
- PMID: 35042807
- PMCID: PMC8795533
- DOI: 10.1073/pnas.2115644119
Darwinian genomics and diversity in the tree of life
Abstract
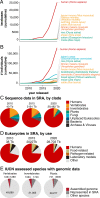
Genomics encompasses the entire tree of life, both extinct and extant, and the evolutionary processes that shape this diversity. To date, genomic research has focused on humans, a small number of agricultural species, and established laboratory models. Fewer than 18,000 of ∼2,000,000 eukaryotic species (<1%) have a representative genome sequence in GenBank, and only a fraction of these have ancillary information on genome structure, genetic variation, gene expression, epigenetic modifications, and population diversity. This imbalance reflects a perception that human studies are paramount in disease research. Yet understanding how genomes work, and how genetic variation shapes phenotypes, requires a broad view that embraces the vast diversity of life. We have the technology to collect massive and exquisitely detailed datasets about the world, but expertise is siloed into distinct fields. A new approach, integrating comparative genomics with cell and evolutionary biology, ecology, archaeology, anthropology, and conservation biology, is essential for understanding and protecting ourselves and our world. Here, we describe potential for scientific discovery when comparative genomics works in close collaboration with a broad range of fields as well as the technical, scientific, and social constraints that must be addressed.
Keywords: biodiversity; comparative genomics; evolution; genomics; natural models.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Kuska B., Beer, Bethesda, and biology: How “genomics” came into being. J. Natl. Cancer Inst. 90, 93 (1998). - PubMed
-
- Fraser C. M., et al. , The minimal gene complement of Mycoplasma genitalium. Science 270, 397–403 (1995). - PubMed
-
- Lander E. S., et al. , International Human Genome Sequencing Consortium, Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). - PubMed
-
- International Chicken Genome Sequencing Consortium, Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004). - PubMed
-
- Lindblad-Toh K., et al. , Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005). - PubMed

