CMG helicase can use ATPγS to unwind DNA: Implications for the rate-limiting step in the reaction mechanism
- PMID: 35042821
- PMCID: PMC8794833
- DOI: 10.1073/pnas.2119580119
CMG helicase can use ATPγS to unwind DNA: Implications for the rate-limiting step in the reaction mechanism
Abstract
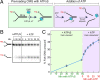
The adenosine triphosphate (ATP) analog ATPγS often greatly slows or prevents enzymatic ATP hydrolysis. The eukaryotic CMG (Cdc45, Mcm2 to 7, GINS) replicative helicase is presumed unable to hydrolyze ATPγS and thus unable to perform DNA unwinding, as documented for certain other helicases. Consequently, ATPγS is often used to "preload" CMG onto forked DNA substrates without unwinding before adding ATP to initiate helicase activity. We find here that CMG does hydrolyze ATPγS and couples it to DNA unwinding. Indeed, the rate of unwinding of a 20- and 30-mer duplex fork of different sequences by CMG is only reduced 1- to 1.5-fold using ATPγS compared with ATP. These findings imply that a conformational change is the rate-limiting step during CMG unwinding, not hydrolysis. Instead of using ATPγS for loading CMG onto DNA, we demonstrate here that nonhydrolyzable adenylyl-imidodiphosphate (AMP-PNP) can be used to preload CMG onto a forked DNA substrate without unwinding.
Keywords: ATPgammaS; CMG helicase; DNA replication; rate-limiting step; staircase model.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

