Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae
- PMID: 35042848
- PMCID: PMC8766457
- DOI: 10.1038/s41467-021-27656-z
Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae
Abstract
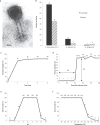
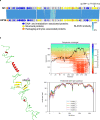
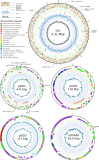
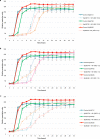
A 30-year-old bombing victim with a fracture-related pandrug-resistant Klebsiella pneumoniae infection after long-term (>700 days) antibiotic therapy is treated with a pre-adapted bacteriophage along with meropenem and colistin, followed by ceftazidime/avibactam. This salvage therapy results in objective clinical, microbiological and radiological improvement of the patient's wounds and overall condition. In support, the bacteriophage and antibiotic combination is highly effective against the patient's K. pneumoniae strain in vitro, in 7-day mature biofilms and in suspensions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

