The R2R3-MYB transcription factor FaMYB63 participates in regulation of eugenol production in strawberry
- PMID: 35043961
- PMCID: PMC8968321
- DOI: 10.1093/plphys/kiac014
The R2R3-MYB transcription factor FaMYB63 participates in regulation of eugenol production in strawberry
Abstract
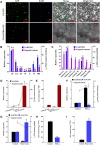
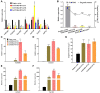
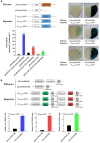
The biosynthetic pathway of volatile phenylpropanoids, including 4-allyl-2-methoxyphenol (eugenol), has been investigated in petunia (Petunia hybrida). However, the regulatory network for eugenol accumulation in strawberry (Fragaria × ananassa Duch.) fruit remains unclear. Here, an R2R3-type MYB transcription factor (TF; FaMYB63) was isolated from strawberry by yeast one-hybrid (Y1H) screening using the promoter of the FaEGS1 (eugenol synthase 1 [EGS 1]) gene, which encodes the enzyme responsible for the last step in eugenol biosynthesis. FaMYB63 is phylogenetically distinct from other R2R3-MYB TFs, including FaEOBІІ (EMISSION OF BENZENOID II [EOBII]), which also participates in regulating eugenol biosynthesis in strawberry receptacles. Reverse transcription quantitative PCR (RT-qPCR) assays showed that the expression of FaMYB63 was tissue-specific and consistent with eugenol content through strawberry fruit development, was repressed by abscisic acid, and was activated by auxins (indole-3-acetic acid). Overexpression and RNA interference-mediated silencing of FaMYB63 resulted in marked changes in the transcript levels of the biosynthetic genes FaEGS1, FaEGS2, and FaCAD1 (cinnamyl alcohol dehydrogenase 1 [CAD1]) and, thereby, the accumulation of eugenol. Electrophoretic mobility shift, Y1H, GUS activity, and dual-luciferase activity assays demonstrated that the transcript levels of FaEOBІІ and FaMYB10 were regulated by FaMYB63, but not the other way around. Together, these results demonstrate that FaMYB63 directly activates FaEGS1, FaEGS2, FaCAD1, FaEOBІІ, and FaMYB10 to induce eugenol biosynthesis during strawberry fruit development. These findings deepen the understanding of the regulatory network that influences eugenol metabolism in an edible fruit crop.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures






Similar articles
-
An R2R3-MYB Transcription Factor Regulates Eugenol Production in Ripe Strawberry Fruit Receptacles.Plant Physiol. 2015 Jun;168(2):598-614. doi: 10.1104/pp.114.252908. Epub 2015 Apr 30. Plant Physiol. 2015. PMID: 25931522 Free PMC article.
-
The fruit-specific transcription factor FaDOF2 regulates the production of eugenol in ripe fruit receptacles.J Exp Bot. 2017 Jul 20;68(16):4529-4543. doi: 10.1093/jxb/erx257. J Exp Bot. 2017. PMID: 28981772
-
The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes.Plant Biotechnol J. 2020 Nov;18(11):2267-2279. doi: 10.1111/pbi.13382. Epub 2020 Apr 13. Plant Biotechnol J. 2020. PMID: 32216018 Free PMC article.
-
Light and abscisic acid independently regulated FaMYB10 in Fragaria × ananassa fruit.Planta. 2015 Apr;241(4):953-65. doi: 10.1007/s00425-014-2228-6. Epub 2014 Dec 23. Planta. 2015. PMID: 25534946
-
Dissecting dietary and semisynthetic volatile phenylpropenes: A compile of their distribution, food properties, health effects, metabolism and toxicities.Crit Rev Food Sci Nutr. 2023;63(32):11105-11124. doi: 10.1080/10408398.2022.2087175. Epub 2022 Jun 16. Crit Rev Food Sci Nutr. 2023. PMID: 35708064 Review.
Cited by
-
Insights into transcription factors controlling strawberry fruit development and ripening.Front Plant Sci. 2022 Oct 10;13:1022369. doi: 10.3389/fpls.2022.1022369. eCollection 2022. Front Plant Sci. 2022. PMID: 36299782 Free PMC article. Review.
-
The Role of PnTCP2 in the Lobed Leaf Formation of Phoebe neurantha var. lobophylla.Int J Mol Sci. 2022 Oct 31;23(21):13296. doi: 10.3390/ijms232113296. Int J Mol Sci. 2022. PMID: 36362084 Free PMC article.
-
Characterization of volatile flavor profiles in three peach cultivars during postharvest storage at various temperatures using HS-SPME-GC-MS.Food Chem X. 2025 May 15;28:102554. doi: 10.1016/j.fochx.2025.102554. eCollection 2025 May. Food Chem X. 2025. PMID: 40497035 Free PMC article.
-
Loss-of-function mutation in anthocyanidin reductase activates the anthocyanin synthesis pathway in strawberry.Mol Hortic. 2024 Sep 14;4(1):33. doi: 10.1186/s43897-024-00106-2. Mol Hortic. 2024. PMID: 39272174 Free PMC article.
-
Colorful hues: insight into the mechanisms of anthocyanin pigmentation in fruit.Plant Physiol. 2023 Jul 3;192(3):1718-1732. doi: 10.1093/plphys/kiad160. Plant Physiol. 2023. PMID: 36913247 Free PMC article.
References
-
- Aharoni A, O’Connell AP (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 - PubMed
-
- Aharoni A, Ric De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

