Expanding the Reach of Precision Oncology by Drugging All KRAS Mutants
- PMID: 35046095
- PMCID: PMC9394389
- DOI: 10.1158/2159-8290.CD-21-1331
Expanding the Reach of Precision Oncology by Drugging All KRAS Mutants
Abstract
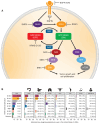
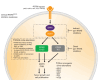
KRAS is the most frequently mutated oncogene, harboring mutations in approximately one in seven cancers. Allele-specific KRASG12C inhibitors are currently changing the treatment paradigm for patients with KRASG12C-mutated non-small cell lung cancer and colorectal cancer. The success of addressing a previously elusive KRAS allele has fueled drug discovery efforts for all KRAS mutants. Pan-KRAS drugs have the potential to address broad patient populations, including KRASG12D-, KRASG12V-, KRASG13D-, KRASG12R-, and KRASG12A-mutant or KRAS wild-type-amplified cancers, as well as cancers with acquired resistance to KRASG12C inhibitors. Here, we review actively pursued allele-specific and pan-KRAS inhibition strategies and their potential utility.
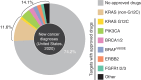
Significance: Mutant-selective KRASG12C inhibitors target a fraction (approximately 13.6%) of all KRAS-driven cancers. A broad arsenal of KRAS drugs is needed to comprehensively conquer KRAS-driven cancers. Conceptually, we foresee two future classes of KRAS medicines: mutant-selective KRAS drugs targeting individual variant alleles and pan-KRAS therapeutics targeting a broad range of KRAS alterations.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal Aet al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

