An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies
- PMID: 35046573
- PMCID: PMC8767531
- DOI: 10.1038/s41591-021-01678-y
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies
Abstract
The emergence of the highly transmissible B.1.1.529 Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is concerning for antibody countermeasure efficacy because of the number of mutations in the spike protein. In this study, we tested a panel of anti-receptor-binding domain monoclonal antibodies (mAbs) corresponding to those in clinical use by Vir Biotechnology (S309, the parent mAb of VIR-7831 (sotrovimab)), AstraZeneca (COV2-2196 and COV2-2130, the parent mAbs of AZD8895 and AZD1061), Regeneron (REGN10933 and REGN10987), Eli Lilly (LY-CoV555 and LY-CoV016) and Celltrion (CT-P59) for their ability to neutralize an infectious B.1.1.529 Omicron isolate. Several mAbs (LY-CoV555, LY-CoV016, REGN10933, REGN10987 and CT-P59) completely lost neutralizing activity against B.1.1.529 virus in both Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells, whereas others were reduced (COV2-2196 and COV2-2130 combination, ~12-fold decrease) or minimally affected (S309). Our results suggest that several, but not all, of the antibodies in clinical use might lose efficacy against the B.1.1.529 Omicron variant.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology and Emergent BioSolutions. J.E.C. has served as a consultant for Luna Biologics and Merck Sharp & Dohme, is on the Scientific Advisory Board of Meissa Vaccines and is the founder of IDBiologics. The Crowe laboratory has received sponsored research agreements from Takeda Vaccines, AstraZeneca and IDBiologics. Vanderbilt University has applied for patents related to two antibodies discussed in this paper. L.A.P. and D.C. are employees of Vir Biotechnology and may hold stock shares in Vir Biotechnology. L.A.P. is a former employee and shareholder in Regeneron Pharmaceuticals. The remaining authors declare no competing interests.
Figures

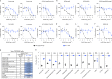
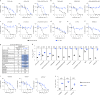
Update of
-
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies.Res Sq [Preprint]. 2021 Dec 27:rs.3.rs-1175516. doi: 10.21203/rs.3.rs-1175516/v1. Res Sq. 2021. Update in: Nat Med. 2022 Mar;28(3):490-495. doi: 10.1038/s41591-021-01678-y. PMID: 34981042 Free PMC article. Updated. Preprint.
References
-
- Tada, T. et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. Preprint at https://www.biorxiv.org/content/10.1101/2021.02.05.430003v1 (2021). - DOI
-
- Wang, P. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature593, 130–135 (2021). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

