Analysis of mRNA and circRNA Expression Profiles of Bovine Monocyte-Derived Macrophages Infected With Mycobacterium avium subsp. paratuberculosis
- PMID: 35046920
- PMCID: PMC8761944
- DOI: 10.3389/fmicb.2021.796922
Analysis of mRNA and circRNA Expression Profiles of Bovine Monocyte-Derived Macrophages Infected With Mycobacterium avium subsp. paratuberculosis
Abstract
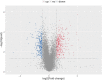
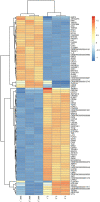
Mycobacterium avium subsp. paratuberculosis (MAP) is the pathogen of Johne's disease (paratuberculosis), which mainly causes chronic infectious granulomatous enteritis in ruminants and has brought huge economic losses to animal husbandry. As a specific intracellular pathogen, when MAP invades the body, it is internalized by macrophages where it is able to replicate by inhibition of the phagosome maturation, escaping the host immune system and surviving, which leads to the spread of the disease. More recent studies have shown that circRNA is involved in many pathological and physiological processes of the body as the molecular sponge of miRNA, the scaffold of RNA binding protein and having the characteristic of being able to translate into protein. In this study, the mRNA and circRNA expression profiles of MAP-infected bovine monocyte-macrophages and uninfected bovine cells were analyzed by high-throughput sequencing. A total of 618 differentially expressed mRNA were screened out, including 322 upregulated mRNA and 296 downregulated mRNA. In addition, the analysis of circRNA differential expression profile showed 39 differentially expressed genes including 12 upregulated and 27 downregulated genes. Moreover, differential genes belonging to cytokine activity, chemokine activity, inflammatory reaction, apoptosis, and other functional groups related to macrophage immune response were significantly enriched in Gene Ontology (GO). Multiple signal pathways including NF-κB, MAPK, Toll-like receptor, IL-17, JAK-STAT, and other signaling pathways related to activating macrophage immune response were significantly enriched in Kyoto Encyclopedia of Genes and Genomes (KEGG). In addition, RT-qPCR technology verified the accuracy of the mRNA sequencing results. In this study, we have obtained the transcriptome information of mRNA and circRNA of bovine monocyte-macrophage infected with MAP. These results will provide data support for the further study of mRNA-miRNA-circRNA network and immune escape mechanism of MAP and will enrich the knowledge of the molecular immune mechanisms of Johne's disease as well.
Keywords: M. avium subsp. paratuberculosis; circRNA; high-throughput sequencing; mRNA; monocyte-macrophage.
Copyright © 2022 Bao, Yao, Wang, Wu, Jiang and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

