SARS-CoV-2 Variants, Vaccines, and Host Immunity
- PMID: 35046961
- PMCID: PMC8761766
- DOI: 10.3389/fimmu.2021.809244
SARS-CoV-2 Variants, Vaccines, and Host Immunity
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new beta coronavirus that emerged at the end of 2019 in the Hubei province of China. SARS-CoV-2 causes coronavirus disease 2019 (COVID-19) and was declared a pandemic by the World Health Organization (WHO) on 11 March 2020. Herd or community immunity has been proposed as a strategy to protect the vulnerable, and can be established through immunity from past infection or vaccination. Whether SARS-CoV-2 infection results in the development of a reservoir of resilient memory cells is under investigation. Vaccines have been developed at an unprecedented rate and 7 408 870 760 vaccine doses have been administered worldwide. Recently emerged SARS-CoV-2 variants are more transmissible with a reduced sensitivity to immune mechanisms. This is due to the presence of amino acid substitutions in the spike protein, which confer a selective advantage. The emergence of variants therefore poses a risk for vaccine effectiveness and long-term immunity, and it is crucial therefore to determine the effectiveness of vaccines against currently circulating variants. Here we review both SARS-CoV-2-induced host immune activation and vaccine-induced immune responses, highlighting the responses of immune memory cells that are key indicators of host immunity. We further discuss how variants emerge and the currently circulating variants of concern (VOC), with particular focus on implications for vaccine effectiveness. Finally, we describe new antibody treatments and future vaccine approaches that will be important as we navigate through the COVID-19 pandemic.
Keywords: SARS-CoV-2; coronavirus; immunity; spike protein; vaccines; variants of concern.
Copyright © 2022 Mistry, Barmania, Mellet, Peta, Strydom, Viljoen, James, Gordon and Pepper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

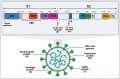

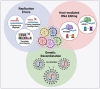

References
-
- WHO . Virtual Press Conference on COVID-19 (2020). Available at: https://www.who.int/docs/default-source/coronaviruse/transcripts/who-aud....
-
- WHO . Novel Coronavirus 2019 (2021). Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

