Automatic Segmentation of Novel Coronavirus Pneumonia Lesions in CT Images Utilizing Deep-Supervised Ensemble Learning Network
- PMID: 35047520
- PMCID: PMC8761973
- DOI: 10.3389/fmed.2021.755309
Automatic Segmentation of Novel Coronavirus Pneumonia Lesions in CT Images Utilizing Deep-Supervised Ensemble Learning Network
Abstract
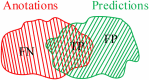
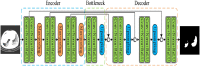
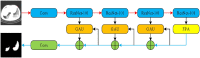
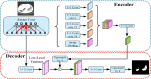
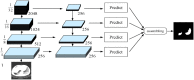
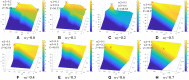
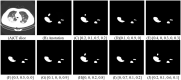
Background: The novel coronavirus disease 2019 (COVID-19) has been spread widely in the world, causing a huge threat to the living environment of people. Objective: Under CT imaging, the structure features of COVID-19 lesions are complicated and varied greatly in different cases. To accurately locate COVID-19 lesions and assist doctors to make the best diagnosis and treatment plan, a deep-supervised ensemble learning network is presented for COVID-19 lesion segmentation in CT images. Methods: Since a large number of COVID-19 CT images and the corresponding lesion annotations are difficult to obtain, a transfer learning strategy is employed to make up for the shortcoming and alleviate the overfitting problem. Based on the reality that traditional single deep learning framework is difficult to extract complicated and varied COVID-19 lesion features effectively that may cause some lesions to be undetected. To overcome the problem, a deep-supervised ensemble learning network is presented to combine with local and global features for COVID-19 lesion segmentation. Results: The performance of the proposed method was validated in experiments with a publicly available dataset. Compared with manual annotations, the proposed method acquired a high intersection over union (IoU) of 0.7279 and a low Hausdorff distance (H) of 92.4604. Conclusion: A deep-supervised ensemble learning network was presented for coronavirus pneumonia lesion segmentation in CT images. The effectiveness of the proposed method was verified by visual inspection and quantitative evaluation. Experimental results indicated that the proposed method has a good performance in COVID-19 lesion segmentation.
Keywords: COVID-19 lesion segmentation; deep learning; deep-supervised ensemble learning network; local and global features; transfer learning; under CT imaging.
Copyright © 2022 Peng, Zhang, Tu and Li.
Conflict of interest statement
HT was employed by company Technique Center, Hunan Great Wall Technology Information Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
A Denoising Self-supervised Approach for COVID-19 Pneumonia Lesion Segmentation with Limited Annotated CT Images.Annu Int Conf IEEE Eng Med Biol Soc. 2021 Nov;2021:3705-3708. doi: 10.1109/EMBC46164.2021.9630215. Annu Int Conf IEEE Eng Med Biol Soc. 2021. PMID: 34892041
-
SSA-Net: Spatial self-attention network for COVID-19 pneumonia infection segmentation with semi-supervised few-shot learning.Med Image Anal. 2022 Jul;79:102459. doi: 10.1016/j.media.2022.102459. Epub 2022 Apr 22. Med Image Anal. 2022. PMID: 35544999 Free PMC article.
-
Lung Lesion Localization of COVID-19 From Chest CT Image: A Novel Weakly Supervised Learning Method.IEEE J Biomed Health Inform. 2021 Jun;25(6):1864-1872. doi: 10.1109/JBHI.2021.3067465. Epub 2021 Jun 3. IEEE J Biomed Health Inform. 2021. PMID: 33739926 Free PMC article.
-
Automatic Segmentation of Multiple Organs on 3D CT Images by Using Deep Learning Approaches.Adv Exp Med Biol. 2020;1213:135-147. doi: 10.1007/978-3-030-33128-3_9. Adv Exp Med Biol. 2020. PMID: 32030668 Review.
-
Machine learning techniques for CT imaging diagnosis of novel coronavirus pneumonia: a review.Neural Comput Appl. 2022 Sep 19:1-19. doi: 10.1007/s00521-022-07709-0. Online ahead of print. Neural Comput Appl. 2022. PMID: 36159188 Free PMC article. Review.
Cited by
-
Identifying the optimal deep learning architecture and parameters for automatic beam aperture definition in 3D radiotherapy.J Appl Clin Med Phys. 2023 Dec;24(12):e14131. doi: 10.1002/acm2.14131. Epub 2023 Sep 5. J Appl Clin Med Phys. 2023. PMID: 37670488 Free PMC article.
References
LinkOut - more resources
Full Text Sources

