Mini review: Recent progress in RT-LAMP enabled COVID-19 detection
- PMID: 35047828
- PMCID: PMC7428436
- DOI: 10.1016/j.snr.2020.100017
Mini review: Recent progress in RT-LAMP enabled COVID-19 detection
Abstract
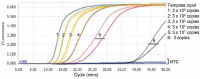
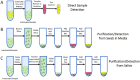
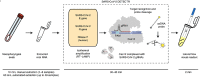
The coronavirus disease 2019 (COVID-19) pandemic has infected millions of people around the globe. The outbreak caused by the novel coronavirus (SARS-CoV-2) poses a great health risk to the public. Therefore, rapid and accurate diagnosis of the virus plays a crucial role in treatment of the disease and saving lives. The current standard method for coronavirus detection is the reverse transcription polymerase chain reaction (RT-PCR) method. However, laboratory-based RT-PCR test for SARS-COV-2 requires complex facilities and elaborate training of operators, thus suffering from limit testing capacity and delayed results. Consequently, isothermal PCR such as loop-mediated isothermal amplification (LAMP) has been emerging as a great alternative to the RT-PCR method. LAMP possesses some fundamental advantages such as amplification at a constant temperature, exclusion of a thermal cycler, a faster test result, and potentially a larger diagnostic capacity, while maintaining similar sensitivity and specificity, thus making it more suitable than the RT-PCR for monitoring a pandemic. Starting with a brief introduction of the working principle of LAMP method, this review summarizes recent progress in LAMP-enabled SARS-CoV-2 viral RNA detection. Lastly, future research directions are discussed. This critical review will motivate biosensor community in furthering the present research, which may pave the road for rapid and large-scale screening of SARS-CoV-2.
Keywords: COVID-19; Isothermal amplification; Molecular diagnosis; RT-LAMP; Rapid.
© 2020 The Author(s).
Conflict of interest statement
There is no Conflict of Interest to declare.
Figures






References
-
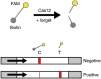
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0513-4. - DOI - PMC - PubMed
-
- Schmid-Burgk J.L., Li D., Feldman D., Slabicki M., Borrajo J., Strecker J., Cleary B., Regev A., Zhang F. LAMP-Seq: population-Scale COVID-19 diagnostics using a compressed barcode space. BioRxiv. 2020 doi: 10.1101/2020.04.06.025635. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
