Apigenin analogues as SARS-CoV-2 main protease inhibitors: In-silico screening approach
- PMID: 35048792
- PMCID: PMC8974217
- DOI: 10.1080/21655979.2022.2027181
Apigenin analogues as SARS-CoV-2 main protease inhibitors: In-silico screening approach
Abstract
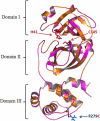
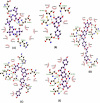
The COVID-19 new variants spread rapidly all over the world, and until now scientists strive to find virus-specific antivirals for its treatment. The main protease of SARS-CoV-2 (Mpro) exhibits high structural and sequence homology to main protease of SARS-CoV (93.23% sequence identity), and their sequence alignment indicated 12 mutated/variant residues. The sequence alignment of SARS-CoV-2 main protease led to identification of only one mutated/variant residue with no significant role in its enzymatic process. Therefore, Mpro was considered as a high-profile drug target in anti-SARS-CoV-2 drug discovery. Apigenin analogues to COVID-19 main protease binding were evaluated. The detailed interactions between the analogues of Apigenin and SARS-CoV-2 Mpro inhibitors were determined as hydrogen bonds, electronic bonds and hydrophobic interactions. The binding energies obtained from the molecular docking of Mpro with Boceprevir, Apigenin, Apigenin 7-glucoside-4'-p-coumarate, Apigenin 7-glucoside-4'-trans-caffeate and Apigenin 7-O-beta-d-glucoside (Cosmosiin) were found to be -6.6, -7.2, -8.8, -8.7 and -8.0 kcal/mol, respectively. Pharmacokinetic parameters and toxicological characteristics obtained by computational techniques and Virtual ADME studies of the Apigenin analogues confirmed that the Apigenin 7-glucoside-4'-p-coumarate is the best candidate for SARS-CoV-2 Mpro inhibition.
Keywords: SARS-Cov-2 main protease; apigenin analogues; docking; inhibitors.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
