TRIM21 suppresses CHK1 activation by preferentially targeting CLASPIN for K63-linked ubiquitination
- PMID: 35048968
- PMCID: PMC8860585
- DOI: 10.1093/nar/gkac011
TRIM21 suppresses CHK1 activation by preferentially targeting CLASPIN for K63-linked ubiquitination
Abstract
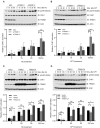
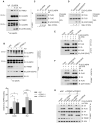
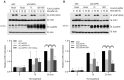
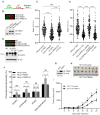
Expression of the E3 ligase TRIM21 is increased in a broad spectrum of cancers; however, the functionally relevant molecular pathway targeted by TRIM21 overexpression remains largely unknown. Here, we show that TRIM21 directly interacts with and ubiquitinates CLASPIN, a mediator for ATR-dependent CHK1 activation. TRIM21-mediated K63-linked ubiquitination of CLASPIN counteracts the K6-linked ubiquitination of CLASPIN which is essential for its interaction with TIPIN and subsequent chromatin loading. We further show that overexpression of TRIM21, but not a TRIM21 catalytically inactive mutant, compromises CHK1 activation, leading to replication fork instability and tumorigenesis. Our findings demonstrate that TRIM21 suppresses CHK1 activation by preferentially targeting CLASPIN for K63-linked ubiquitination, providing a potential target for cancer therapy.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Regulation of ATR-CHK1 signaling by ubiquitination of CLASPIN.Biochem Soc Trans. 2022 Oct 31;50(5):1471-1480. doi: 10.1042/BST20220729. Biochem Soc Trans. 2022. PMID: 36196914 Review.
-
Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts.Cell Cycle. 2013 Jan 15;12(2):332-45. doi: 10.4161/cc.23177. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255133 Free PMC article.
-
Claspin - checkpoint adaptor and DNA replication factor.FEBS J. 2019 Feb;286(3):441-455. doi: 10.1111/febs.14594. Epub 2018 Jun 29. FEBS J. 2019. PMID: 29931808 Review.
-
Chk1 and Claspin potentiate PCNA ubiquitination.Genes Dev. 2008 May 1;22(9):1147-52. doi: 10.1101/gad.1632808. Genes Dev. 2008. PMID: 18451105 Free PMC article.
-
Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response.Mol Biol Cell. 2005 Nov;16(11):5269-82. doi: 10.1091/mbc.e05-07-0671. Epub 2005 Sep 7. Mol Biol Cell. 2005. PMID: 16148040 Free PMC article.
Cited by
-
TRIM21 Promotes Endothelial Cell Activation via Accelerating SOCS3 Ubiquitination Degradation in Atherosclerosis.Cardiovasc Toxicol. 2025 Mar;25(3):395-410. doi: 10.1007/s12012-025-09965-7. Epub 2025 Feb 8. Cardiovasc Toxicol. 2025. PMID: 39921798
-
METTL21A promotes hepatocellular carcinoma progression via methylating and stabilizing BAG3.NPJ Precis Oncol. 2025 Jul 10;9(1):234. doi: 10.1038/s41698-025-01021-5. NPJ Precis Oncol. 2025. PMID: 40640484 Free PMC article.
-
TRIM21-Promoted FSP1 Plasma Membrane Translocation Confers Ferroptosis Resistance in Human Cancers.Adv Sci (Weinh). 2023 Oct;10(29):e2302318. doi: 10.1002/advs.202302318. Epub 2023 Aug 16. Adv Sci (Weinh). 2023. PMID: 37587773 Free PMC article.
-
CDC7 Inhibition Potentiates Antitumor Efficacy of PARP Inhibitor in Advanced Ovarian Cancer.Adv Sci (Weinh). 2024 Dec;11(45):e2403782. doi: 10.1002/advs.202403782. Epub 2024 Oct 16. Adv Sci (Weinh). 2024. PMID: 39412086 Free PMC article.
-
Involvement of the ubiquitin-proteasome system in the regulation of the tumor microenvironment and progression.Genes Dis. 2024 Feb 2;12(2):101240. doi: 10.1016/j.gendis.2024.101240. eCollection 2025 Mar. Genes Dis. 2024. PMID: 39759114 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

