Slt2-MAPK/RNS1 Controls Conidiation via Direct Regulation of the Central Regulatory Pathway in the Fungus Metarhizium robertsii
- PMID: 35049966
- PMCID: PMC8779605
- DOI: 10.3390/jof8010026
Slt2-MAPK/RNS1 Controls Conidiation via Direct Regulation of the Central Regulatory Pathway in the Fungus Metarhizium robertsii
Abstract
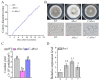
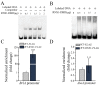
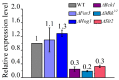
Ascomycete fungi usually produce small hydrophobic asexual conidia that are easily dispersed and essential for long-term survival under a variety of environmental conditions. Several upstream signaling regulators have been documented to control conidiation via regulation of the central regulatory pathway that contains the transcription factors BrlA, AbaA and WetA. Here, we showed that the Slt2-MAPK signaling pathway and the transcription factor RNS1 constitute a novel upstream signaling cascade that activates the central regulatory pathway for conidiation in the Ascomycetes fungus M. robertsii. The BrlA gene has two overlapping transcripts BrlAα and BrlAβ; they have the same major ORF, but the 5' UTR of BrlAβ is 835 bp longer than the one of BrlAα. During conidiation, Slt2 phosphorylates the serine residue at the position 306 in RNS1, which self-induces. RNS1 binds to the BM2 motif in the promoter of the BrlA gene and induces the expression of the transcript BlrAα, which in turn activates the expression of the genes AbaA and WetA. In conclusion, the Slt2/RNS1 cascade represents a novel upstream signaling pathway that initiates conidiation via direct activation of the central regulatory pathway. This work provides significant mechanistic insights into the production of asexual conidia in an Ascomycete fungus.
Keywords: BrlA; MAPK; Metarhizium; RNS1; conidiation; fungi; phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Ebbole D.J. Cellular and Molecular Biology of Filamentous Fungi. ASM Press; Washington, DC, USA: 2010. The conidium; pp. 577–590.
Grants and funding
LinkOut - more resources
Full Text Sources

