Genomic Characterization of Parengyodontium torokii sp. nov., a Biofilm-Forming Fungus Isolated from Mars 2020 Assembly Facility
- PMID: 35050006
- PMCID: PMC8778116
- DOI: 10.3390/jof8010066
Genomic Characterization of Parengyodontium torokii sp. nov., a Biofilm-Forming Fungus Isolated from Mars 2020 Assembly Facility
Abstract
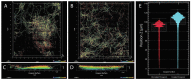
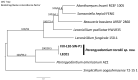
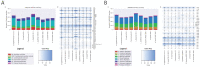
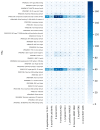
A fungal strain (FJII-L10-SW-P1) was isolated from the Mars 2020 spacecraft assembly facility and exhibited biofilm formation on spacecraft-qualified Teflon surfaces. The reconstruction of a six-loci gene tree (ITS, LSU, SSU, RPB1 and RPB2, and TEF1) using multi-locus sequence typing (MLST) analyses of the strain FJII-L10-SW-P1 supported a close relationship to other known Parengyodontium album subclade 3 isolates while being phylogenetically distinct from subclade 1 strains. The zig-zag rachides morphology of the conidiogenous cells and spindle-shaped conidia were the distinct morphological characteristics of the P. album subclade 3 strains. The MLST data and morphological analysis supported the conclusion that the P. album subclade 3 strains could be classified as a new species of the genus Parengyodontium and placed in the family Cordycipitaceae. The name Parengyodontium torokii sp. nov. is proposed to accommodate the strain, with FJII-L10-SW-P1 as the holotype. The genome of the FJII-L10-SW-P1 strain was sequenced, annotated, and the secondary metabolite clusters were identified. Genes predicted to be responsible for biofilm formation and adhesion to surfaces were identified. Homology-based assignment of gene ontologies to the predicted proteome of P. torokii revealed the presence of gene clusters responsible for synthesizing several metabolic compounds, including a cytochalasin that was also verified using traditional metabolomic analysis.
Keywords: biofilm; fungi; genomics; mars 2020 mission; metabolomics; morphological analysis; phylogenetic analysis.
Conflict of interest statement
The authors declare that there are no conflict of interest. This manuscript was prepared as an account of work sponsored by NASA, an agency of the US Government. The US Government, NASA, California Institute of Technology, Jet Propulsion Laboratory, and their employees make no warranty, expressed or implied, or assume any liability or responsibility for the accuracy, completeness, or usefulness of information, apparatus, product, or process disclosed in this manuscript, or represents that its use would not infringe upon privately held rights. The use of, and references to any commercial product, process, or service does not necessarily constitute or imply endorsement, recommendation, or favoring by the US Government, NASA, California Institute of Technology, or Jet Propulsion Laboratory. Views and opinions presented herein by the authors of this manuscript do not necessarily reflect those of the US Government, NASA, California Institute of Technology, or Jet Propulsion Laboratory, and shall not be used for advertisements or product endorsements.
Figures


Similar articles
-
Description and Genome Characterization of Three Novel Fungal Strains Isolated from Mars 2020 Mission-Associated Spacecraft Assembly Facility Surfaces-Recommendations for Two New Genera and One Species.J Fungi (Basel). 2022 Dec 23;9(1):31. doi: 10.3390/jof9010031. J Fungi (Basel). 2022. PMID: 36675851 Free PMC article.
-
Genomic characterization and radiation tolerance of Naganishia kalamii sp. nov. and Cystobasidium onofrii sp. nov. from Mars 2020 mission assembly facilities.IMA Fungus. 2023 Aug 11;14(1):15. doi: 10.1186/s43008-023-00119-4. IMA Fungus. 2023. PMID: 37568226 Free PMC article.
-
Diversity of Parengyodontium spp. strains isolated from the cultural heritage environment: Phylogenetic diversity, phenotypical diversity, and occurrence.Mycologia. 2022 Sep-Oct;114(5):825-840. doi: 10.1080/00275514.2022.2094046. Epub 2022 Jul 29. Mycologia. 2022. PMID: 35904483
-
Cutaneous hyalohyphomycosis due to Parengyodontium album gen. et comb. nov.Med Mycol. 2016 Oct 1;54(7):699-713. doi: 10.1093/mmy/myw025. Epub 2016 May 9. Med Mycol. 2016. PMID: 27161787
-
Research progress on ecological adaptation and prevention of Parengyodontium album on the surface of cultural relics.Ying Yong Sheng Tai Xue Bao. 2024 Mar 18;35(3):837-846. doi: 10.13287/j.1001-9332.202403.031. Ying Yong Sheng Tai Xue Bao. 2024. PMID: 38646772 Review. English.
Cited by
-
Description and Genome Characterization of Three Novel Fungal Strains Isolated from Mars 2020 Mission-Associated Spacecraft Assembly Facility Surfaces-Recommendations for Two New Genera and One Species.J Fungi (Basel). 2022 Dec 23;9(1):31. doi: 10.3390/jof9010031. J Fungi (Basel). 2022. PMID: 36675851 Free PMC article.
-
Genomic characterization and radiation tolerance of Naganishia kalamii sp. nov. and Cystobasidium onofrii sp. nov. from Mars 2020 mission assembly facilities.IMA Fungus. 2023 Aug 11;14(1):15. doi: 10.1186/s43008-023-00119-4. IMA Fungus. 2023. PMID: 37568226 Free PMC article.
-
Genomic and morphological characterization of Knufia obscura isolated from the Mars 2020 spacecraft assembly facility.Sci Rep. 2024 May 28;14(1):12249. doi: 10.1038/s41598-024-61115-1. Sci Rep. 2024. PMID: 38806503 Free PMC article.
-
Understanding the Complexities and Changes of the Astronaut Microbiome for Successful Long-Duration Space Missions.Life (Basel). 2022 Mar 28;12(4):495. doi: 10.3390/life12040495. Life (Basel). 2022. PMID: 35454986 Free PMC article. Review.
References
-
- Blachowicz A., Chiang A.J., Elsaesser A., Kalkum M., Ehrenfreund P., Stajich J.E., Torok T., Wang C.C.C., Venkateswaran K. Proteomic and Metabolomic Characteristics of Extremophilic Fungi under Simulated Mars Conditions. Front. Microbiol. 2019;10:1013. doi: 10.3389/fmicb.2019.01013. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

