Comparative Genomics, Phylogenetics, Biogeography, and Effects of Climate Change on Toddalia asiatica (L.) Lam. (Rutaceae) from Africa and Asia
- PMID: 35050119
- PMCID: PMC8781850
- DOI: 10.3390/plants11020231
Comparative Genomics, Phylogenetics, Biogeography, and Effects of Climate Change on Toddalia asiatica (L.) Lam. (Rutaceae) from Africa and Asia
Abstract
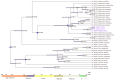
In the present study, two samples of Toddalia asiatica species, both collected from Kenya, were sequenced and comparison of their genome structures carried out with T. asiatica species from China, available in the NCBI database. The genome size of both species from Africa was 158, 508 base pairs, which was slightly larger, compared to the reference genome of T. asiatica from Asia (158, 434 bp). The number of genes was 113 for both species from Africa, consisting of 79 protein-coding genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Toddalia asiatica from Asia had 115 genes with 81 protein-coding genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Both species compared displayed high similarity in gene arrangement. The gene number, orientation, and order were highly conserved. The IR/SC boundary structures were the same in all chloroplast genomes. A comparison of pairwise sequences indicated that the three regions (trnH-psbA, rpoB, and ycf1) were more divergent and can be useful in developing effective genetic markers. Phylogenetic analyses of the complete cp genomes and 79 protein-coding genes indicated that the Toddalia species collected from Africa were sister to T. asiatica collected from Asia. Both species formed a sister clade to the Southwest Pacific and East Asian species of Zanthoxylum. These results supported the previous studies of merging the genus Toddalia with Zanthoxylum and taxonomic change of Toddalia asiatica to Zanthoxylum asiaticum, which should also apply for the African species of Toddalia. Biogeographic results demonstrated that the two samples of Toddalia species from Africa diverged from T. asiatica from Asia (3.422 Mya, 95% HPD). These results supported an Asian origin of Toddalia species and later dispersal to Africa and Madagascar. The maxent model analysis showed that Asia would have an expansion of favorable areas for Toddalia species in the future. In Africa, there will be contraction and expansion of the favorable areas for the species. The availability of these cp genomes will provide valuable genetic resources for further population genetics and biogeographic studies of these species. However, more T. asiatica species collected from a wide geographical range are required.
Keywords: Rutaceae; biogeography; comparative analysis; divergent hotspots; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Kubitzki K. Flowering Plants. Eudicots: Sapindales, Cucurbitales, Myrtaceae. Volume 10 Springer Science & Business Media; Amtsgericht Bonn, Germany: 2010.
-
- Groppo M., Kallunki J.A., Pirani J.R., Antonelli A. Chilean Pitavia more closely related to Oceania and Old World Rutaceae than to Neotropical groups: Evidence from two cpDNA non-coding regions, with a new subfamilial classification of the family. PhytoKeys. 2012;9:9–29. doi: 10.3897/phytokeys.19.3912. - DOI - PMC - PubMed
-
- Chase M.W., Christenhusz M., Fay M., Byng J., Judd W.S., Soltis D., Mabberley D., Sennikov A., Soltis P.S., Stevens P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

