Interplay of vascular endothelial growth factor receptors in organ-specific vessel maintenance
- PMID: 35050301
- PMCID: PMC8785977
- DOI: 10.1084/jem.20210565
Interplay of vascular endothelial growth factor receptors in organ-specific vessel maintenance
Abstract
Vascular endothelial growth factors (VEGFs) and their receptors (VEGFRs) are quintessential for the development and maintenance of blood and lymphatic vessels. However, genetic interactions between the VEGFRs are poorly understood. VEGFR2 is the dominant receptor that is required for the growth and survival of the endothelium, whereas deletion of VEGFR1 or VEGFR3 was reported to induce vasculature overgrowth. Here we show that vascular regression induced by VEGFR2 deletion in postnatal and adult mice is aggravated by additional deletion of VEGFR1 or VEGFR3 in the intestine, kidney, and pancreas, but not in the liver or kidney glomeruli. In the adult mice, hepatic and intestinal vessels regressed within a few days after gene deletion, whereas vessels in skin and retina remained stable for at least four weeks. Our results show changes in endothelial transcriptomes and organ-specific vessel maintenance mechanisms that are dependent on VEGFR signaling pathways and reveal previously unknown functions of VEGFR1 and VEGFR3 in endothelial cells.
© 2022 Karaman et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures
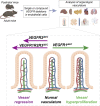









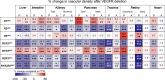




Comment in
-
Vascular heterogeneity: VEGF receptors make blood vessels special.J Exp Med. 2022 Mar 7;219(3):e20212539. doi: 10.1084/jem.20212539. Epub 2022 Jan 27. J Exp Med. 2022. PMID: 35085381 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

