Immune landscape of human placental villi using single-cell analysis
- PMID: 35050308
- PMCID: PMC8935213
- DOI: 10.1242/dev.200013
Immune landscape of human placental villi using single-cell analysis
Abstract
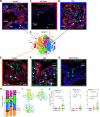
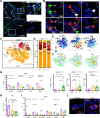
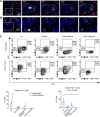
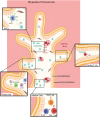
Maintenance of a healthy pregnancy is reliant on a successful balance between the fetal and maternal immune systems. Although the maternal mechanisms responsible have been well studied, those used by the fetal immune system remain poorly understood. Using suspension mass cytometry and various imaging modalities, we report a complex immune system within the mid-gestation (17-23 weeks) human placental villi (PV). Consistent with recent reports in other fetal organs, T cells with memory phenotypes, although rare in abundance, were detected within the PV tissue and vasculature. Moreover, we determined that T cells isolated from PV samples may be more proliferative after T cell receptor stimulation than adult T cells at baseline. Collectively, we identified multiple subtypes of fetal immune cells within the PV and specifically highlight the enhanced proliferative capacity of fetal PV T cells.
Keywords: Immune cells; Placenta; Pregnancy; T cells.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

