Leukocytes mediate disease pathogenesis in the Ndufs4(KO) mouse model of Leigh syndrome
- PMID: 35050903
- PMCID: PMC8983133
- DOI: 10.1172/jci.insight.156522
Leukocytes mediate disease pathogenesis in the Ndufs4(KO) mouse model of Leigh syndrome
Abstract
Symmetric, progressive, necrotizing lesions in the brainstem are a defining feature of Leigh syndrome (LS). A mechanistic understanding of the pathogenesis of these lesions has been elusive. Here, we report that leukocyte proliferation is causally involved in the pathogenesis of LS. Depleting leukocytes with a colony-stimulating factor 1 receptor inhibitor disrupted disease progression, including suppression of CNS lesion formation and a substantial extension of survival. Leukocyte depletion rescued diverse symptoms, including seizures, respiratory center function, hyperlactemia, and neurologic sequelae. These data reveal a mechanistic explanation for the beneficial effects of mTOR inhibition. More importantly, these findings dramatically alter our understanding of the pathogenesis of LS, demonstrating that immune involvement is causal in disease. This work has important implications for the mechanisms of mitochondrial disease and may lead to novel therapeutic strategies.
Keywords: Genetic diseases; Inflammation; Mitochondria; Mouse models; Neuroscience.
Figures
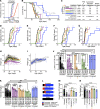
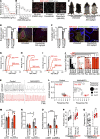
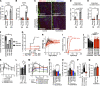

Similar articles
-
Disruption of adaptive immunity does not attenuate disease in the Ndufs4(-/-) model of Leigh syndrome.PLoS One. 2025 Jun 10;20(6):e0324268. doi: 10.1371/journal.pone.0324268. eCollection 2025. PLoS One. 2025. PMID: 40493653 Free PMC article.
-
Peripheral macrophages drive CNS disease in the Ndufs4(-/-) model of Leigh syndrome.Brain Pathol. 2023 Nov;33(6):e13192. doi: 10.1111/bpa.13192. Epub 2023 Aug 8. Brain Pathol. 2023. PMID: 37552802 Free PMC article.
-
Interferon-gamma contributes to disease progression in the Ndufs4(-/-) model of Leigh syndrome.Neuropathol Appl Neurobiol. 2024 Jun;50(3):e12977. doi: 10.1111/nan.12977. Neuropathol Appl Neurobiol. 2024. PMID: 38680020
-
Ndufs4 knockout mouse models of Leigh syndrome: pathophysiology and intervention.Brain. 2022 Mar 29;145(1):45-63. doi: 10.1093/brain/awab426. Brain. 2022. PMID: 34849584 Free PMC article. Review.
-
Disease models of Leigh syndrome: From yeast to organoids.J Inherit Metab Dis. 2024 Nov;47(6):1292-1321. doi: 10.1002/jimd.12804. Epub 2024 Oct 9. J Inherit Metab Dis. 2024. PMID: 39385390 Free PMC article. Review.
Cited by
-
Activated microglia and neuroinflammation as a pathogenic mechanism in Leigh syndrome.Front Neurosci. 2023 Jan 18;16:1068498. doi: 10.3389/fnins.2022.1068498. eCollection 2022. Front Neurosci. 2023. PMID: 36741056 Free PMC article.
-
Disruption of adaptive immunity does not attenuate disease in the Ndufs4(-/-) model of Leigh syndrome.PLoS One. 2025 Jun 10;20(6):e0324268. doi: 10.1371/journal.pone.0324268. eCollection 2025. PLoS One. 2025. PMID: 40493653 Free PMC article.
-
Anti-AQP4-IgG-positive Leigh syndrome: A case report and review of the literature.Front Pediatr. 2023 Feb 6;11:1046731. doi: 10.3389/fped.2023.1046731. eCollection 2023. Front Pediatr. 2023. PMID: 36814591 Free PMC article.
-
Caspase-11 drives macrophage hyperinflammation in models of Polg-related mitochondrial disease.bioRxiv [Preprint]. 2025 Jan 17:2024.05.11.593693. doi: 10.1101/2024.05.11.593693. bioRxiv. 2025. Update in: Nat Commun. 2025 May 20;16(1):4640. doi: 10.1038/s41467-025-59907-8. PMID: 38798587 Free PMC article. Updated. Preprint.
-
Neurological Phenotypes in Mouse Models of Mitochondrial Disease and Relevance to Human Neuropathology.Int J Mol Sci. 2023 Jun 2;24(11):9698. doi: 10.3390/ijms24119698. Int J Mol Sci. 2023. PMID: 37298649 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

