Use of Raman and Raman optical activity to extract atomistic details of saccharides in aqueous solution
- PMID: 35051172
- PMCID: PMC8806073
- DOI: 10.1371/journal.pcbi.1009678
Use of Raman and Raman optical activity to extract atomistic details of saccharides in aqueous solution
Abstract
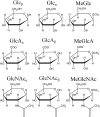
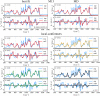
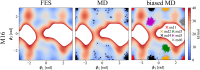
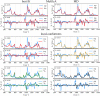
Sugars are crucial components in biosystems and industrial applications. In aqueous environments, the natural state of short saccharides or charged glycosaminoglycans is floating and wiggling in solution. Therefore, tools to characterize their structure in a native aqueous environment are crucial but not always available. Here, we show that a combination of Raman/ROA and, on occasions, NMR experiments with Molecular Dynamics (MD) and Quantum Mechanics (QM) is a viable method to gain insights into structural features of sugars in solutions. Combining these methods provides information about accessible ring puckering conformers and their proportions. It also provides information about the conformation of the linkage between the sugar monomers, i.e., glycosidic bonds, allowing for identifying significantly accessible conformers and their relative abundance. For mixtures of sugar moieties, this method enables the deconvolution of the Raman/ROA spectra to find the actual amounts of its molecular constituents, serving as an effective analytical technique. For example, it allows calculating anomeric ratios for reducing sugars and analyzing more complex sugar mixtures to elucidate their real content. Altogether, we show that combining Raman/ROA spectroscopies with simulations is a versatile method applicable to saccharides. It allows for accessing many features with precision comparable to other methods routinely used for this task, making it a viable alternative. Furthermore, we prove that the proposed technique can scale up by studying the complicated raffinose trisaccharide, and therefore, we expect its wide adoption to characterize sugar structural features in solution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




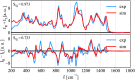
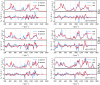
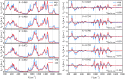


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

