Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater
- PMID: 35051459
- PMCID: PMC8762835
- DOI: 10.1016/j.scitotenv.2022.153171
Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater
Abstract
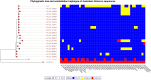
On the 26th of November 2021, the World Health Organization (WHO) designated the newly detected B.1.1.529 lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) the Omicron Variant of Concern (VOC). The genome of the Omicron VOC contains more than 50 mutations, many of which have been associated with increased transmissibility, differing disease severity, and potential to evade immune responses developed for previous VOCs such as Alpha and Delta. In the days since the designation of B.1.1.529 as a VOC, infections with the lineage have been reported in countries around the globe and many countries have implemented travel restrictions and increased border controls in response. We putatively detected the Omicron variant in an aircraft wastewater sample from a flight arriving to Darwin, Australia from Johannesburg, South Africa on the 25th of November 2021 via positive results on the CDC N1, CDC N2, and del(69-70) RT-qPCR assays per guidance from the WHO. The Australian Northern Territory Health Department detected one passenger onboard the flight who was infected with SARS-CoV-2, which was determined to be the Omicron VOC by sequencing of a nasopharyngeal swab sample. Subsequent sequencing of the aircraft wastewater sample using the ARTIC V3 protocol with Nanopore and ATOPlex confirmed the presence of the Omicron variant with a consensus genome that clustered with the B.1.1.529 BA.1 sub-lineage. Our detection and confirmation of a single onboard Omicron infection via aircraft wastewater further bolsters the important role that aircraft wastewater can play as an independent and unintrusive surveillance point for infectious diseases, particularly coronavirus disease 2019.
Keywords: Aircraft; COVID-19; Enveloped viruses; Human health risks; SARS-CoV-2; Wastewater surveillance.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J Travel Med. 2020;27 - PMC - PubMed
-
- Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773.
-
- Besselsen D.G., Wagner A.M., Loganbill J.K. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp Med. 2002;52:111–116. - PubMed
-
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

