Ancient DNA Methods Improve Forensic DNA Profiling of Korean War and World War II Unknowns
- PMID: 35052469
- PMCID: PMC8774965
- DOI: 10.3390/genes13010129
Ancient DNA Methods Improve Forensic DNA Profiling of Korean War and World War II Unknowns
Abstract
The integration of massively parallel sequencing (MPS) technology into forensic casework has been of particular benefit to the identification of unknown military service members. However, highly degraded or chemically treated skeletal remains often fail to provide usable DNA profiles, even with sensitive mitochondrial (mt) DNA capture and MPS methods. In parallel, the ancient DNA field has developed workflows specifically for degraded DNA, resulting in the successful recovery of nuclear DNA and mtDNA from skeletal remains as well as sediment over 100,000 years old. In this study we use a set of disinterred skeletal remains from the Korean War and World War II to test if ancient DNA extraction and library preparation methods improve forensic DNA profiling. We identified an ancient DNA extraction protocol that resulted in the recovery of significantly more human mtDNA fragments than protocols previously used in casework. In addition, utilizing single-stranded rather than double-stranded library preparation resulted in increased attainment of reportable mtDNA profiles. This study emphasizes that the combination of ancient DNA extraction and library preparation methods evaluated here increases the success rate of DNA profiling, and likelihood of identifying historical remains.
Keywords: ancient DNA; degraded DNA; forensic DNA profiling; human identification; massively parallel sequencing (MPS); mitochondrial DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

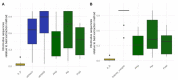

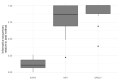

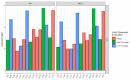
References
-
- First Criminal Conviction Secured with Next-Gen Forensic DNA Technology. 2019. [(accessed on 2 December 2021)]. Available online: https://verogen.com/first-criminal-conviction-with-next-gen-forensic-dna/
-
- National DNA Index System (NDIS) Operational Procedures Manual. FBI Laboratory; Quantico, VA, USA: 2021. Version 10.
-
- Marshall C., Andreaggi K., Daniels-Higginbotham J., Oliver R.S., Barritt-Ross S., McMahon T.P. Performance evaluation of a mitogenome capture and Illumina sequencing protocol using non-probative, case-type skeletal samples: Implications for the use of a positive control in a next-generation sequencing procedure. Forensic Sci. Int. Genet. 2017;31:198–206. doi: 10.1016/j.fsigen.2017.09.001. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

