Wnt Pathway Extracellular Components and Their Essential Roles in Bone Homeostasis
- PMID: 35052478
- PMCID: PMC8775112
- DOI: 10.3390/genes13010138
Wnt Pathway Extracellular Components and Their Essential Roles in Bone Homeostasis
Abstract
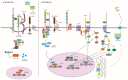
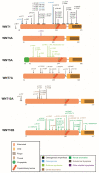
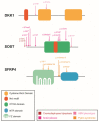
The Wnt pathway is involved in several processes essential for bone development and homeostasis. For proper functioning, the Wnt pathway is tightly regulated by numerous extracellular elements that act by both activating and inhibiting the pathway at different moments. This review aims to describe, summarize and update the findings regarding the extracellular modulators of the Wnt pathway, including co-receptors, ligands and inhibitors, in relation to bone homeostasis, with an emphasis on the animal models generated, the diseases associated with each gene and the bone processes in which each member is involved. The precise knowledge of all these elements will help us to identify possible targets that can be used as a therapeutic target for the treatment of bone diseases such as osteoporosis.
Keywords: Wnt pathway; bone; co-receptors; inhibitors; ligands.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

