Nonsense-Mediated mRNA Decay, a Finely Regulated Mechanism
- PMID: 35052820
- PMCID: PMC8773229
- DOI: 10.3390/biomedicines10010141
Nonsense-Mediated mRNA Decay, a Finely Regulated Mechanism
Abstract
Nonsense-mediated mRNA decay (NMD) is both a mechanism for rapidly eliminating mRNAs carrying a premature termination codon and a pathway that regulates many genes. This implies that NMD must be subject to regulation in order to allow, under certain physiological conditions, the expression of genes that are normally repressed by NMD. Therapeutically, it might be interesting to express certain NMD-repressed genes or to allow the synthesis of functional truncated proteins. Developing such approaches will require a good understanding of NMD regulation. This review describes the different levels of this regulation in human cells.
Keywords: UPF proteins; gene expression; nonsense-mediated mRNA decay; regulation; therapeutic perspectives.
Conflict of interest statement
Author declares no conflicts of interest.
Figures
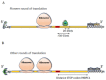
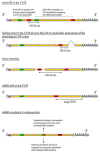
References
Publication types
LinkOut - more resources
Full Text Sources

