Breeding for Economically and Environmentally Sustainable Wheat Varieties: An Integrated Approach from Genomics to Selection
- PMID: 35053148
- PMCID: PMC8773325
- DOI: 10.3390/biology11010149
Breeding for Economically and Environmentally Sustainable Wheat Varieties: An Integrated Approach from Genomics to Selection
Abstract
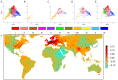
There is currently a strong societal demand for sustainability, quality, and safety in bread wheat production. To address these challenges, new and innovative knowledge, resources, tools, and methods to facilitate breeding are needed. This starts with the development of high throughput genomic tools including single nucleotide polymorphism (SNP) arrays, high density molecular marker maps, and full genome sequences. Such powerful tools are essential to perform genome-wide association studies (GWAS), to implement genomic and phenomic selection, and to characterize the worldwide diversity. This is also useful to breeders to broaden the genetic basis of elite varieties through the introduction of novel sources of genetic diversity. Improvement in varieties particularly relies on the detection of genomic regions involved in agronomical traits including tolerance to biotic (diseases and pests) and abiotic (drought, nutrient deficiency, high temperature) stresses. When enough resolution is achieved, this can result in the identification of candidate genes that could further be characterized to identify relevant alleles. Breeding must also now be approached through in silico modeling to simulate plant development, investigate genotype × environment interactions, and introduce marker-trait linkage information in the models to better implement genomic selection. Breeders must be aware of new developments and the information must be made available to the world wheat community to develop new high-yielding varieties that can meet the challenge of higher wheat production in a sustainable and fluctuating agricultural context. In this review, we compiled all knowledge and tools produced during the BREEDWHEAT project to show how they may contribute to face this challenge in the coming years.
Keywords: Triticum aestivum; diversity; genomic selection; high throughput phenotyping; molecular tools; wheat; wheat breeding; wheat database.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
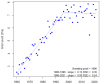
- Brisson N., Gate G., Gouache D., Charmet G., Oury F.-X., Huard F. Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crops Res. 2010;119:201–212. doi: 10.1016/j.fcr.2010.07.012. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

